Frequency of Fecal Carriage of ESBL Resistance Genes in Multidrug-Resistant Pseudomonas aeruginosa Isolates from Cancer Patients at Laquintinie Hospital, Douala, Littoral Region, Cameroon
- PMID: 38962396
- PMCID: PMC11222006
- DOI: 10.1155/2024/7685878
Frequency of Fecal Carriage of ESBL Resistance Genes in Multidrug-Resistant Pseudomonas aeruginosa Isolates from Cancer Patients at Laquintinie Hospital, Douala, Littoral Region, Cameroon
Abstract
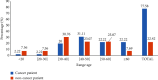
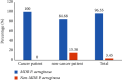
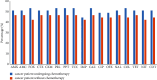
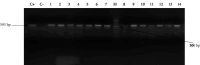
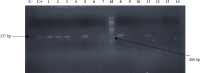
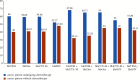
Background. Opportunistic infections are the second cause of death among cancer patients. This study aimed at determining the antimicrobial profile and the prevalence of extended-spectrum beta-lactamase (ESBL)-gene carriage of Pseudomonas aeruginosa isolates among cancer patients at the Douala Laquintinie Hospital, Littoral Region of Cameroon. Between October 2021 and March 2023, 507 study participants were recruited among whom 307 (60.55%) were cancer patients, compared to 200 (39.45%) noncancer patients. Fifty-eight P. aeruginosa isolates were isolated from fecal samples of forty-five cancer patients and thirteen noncancer patients using Cetrimide agar. The antimicrobial resistance profile of the isolates was determined using the Kirby-Bauer disk diffusion method. The polymerase chain reaction was used to detect the presence of extended-spectrum beta-lactamase genes among P. aeruginosa isolates. P. aeruginosa showed significant resistance rates in cancer patients compared to noncancer patients to imipenem, cefotaxime, and ceftazidime, piperacillin-tazobactam, ticarcillin-clavulanic acid, and ciprofloxacin. The multidrug resistance (MDR) rate was significantly (p < 0.05) higher in cancer patients than in noncancer patients. The frequency of beta-lactamase genes in the 58 ESBL-producing P. aeruginosa isolates was determined as 72.41% for bla TEM, 37.93% for bla OXA, 74.14% for blaCTX-M, and 44.83% for bla SHV genes. The study revealed an alarmingly high prevalence of fecal carriage of ESBL-producing P. aeruginosa with a high rate of MDR among cancer patients. It indicates that regular monitoring and surveillance of ESBL-producing P. aeruginosa among cancer patients are needed to improve the management of patients.
Copyright © 2024 Michael F. Kengne et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
Antibiotic Resistance Profile of Enterovirulent E. coli Isolates Harboring Broad-Spectrum Beta-Lactamase Genes in Cancer Patients at the Laquintinie Hospital in Douala, Littoral Region, Cameroon.Int J Microbiol. 2025 Jan 9;2025:4224807. doi: 10.1155/ijm/4224807. eCollection 2025. Int J Microbiol. 2025. PMID: 39822382 Free PMC article.
-
Prevalence of ESBL resistance genes and fecal carriage of multidrug-resistant Pseudomonas aeruginosa isolates from patients with chronic kidney disease at the Laquintinie Hospital in Douala, Littoral Region, Cameroon.J Infect Public Health. 2025 Feb;18(2):102651. doi: 10.1016/j.jiph.2024.102651. Epub 2025 Jan 6. J Infect Public Health. 2025. PMID: 39787731
-
Detection of Extended-Spectrum β-Lactamases, Metallo-β-Lactamases, Antimicrobial Resistance Profiles, and Biofilm-Forming Capacity in Pseudomonas aeruginosa Strains Recovered From Dogs With Otitis Externa in Italy.Vet Med Int. 2025 May 16;2025:5566151. doi: 10.1155/vmi/5566151. eCollection 2025. Vet Med Int. 2025. PMID: 40415870 Free PMC article.
-
[Investigation of beta-lactamase genes and clonal relationship among the extended-spectrum beta-lactamase producing nosocomial Escherichia coli isolates].Mikrobiyol Bul. 2015 Jan;49(1):15-25. doi: 10.5578/mb.8437. Mikrobiyol Bul. 2015. PMID: 25706727 Turkish.
-
Molecular detection of Metallo-Beta-Lactamase and alginate in multidrug resistance Pseudomonas aeruginosa isolated from the clinical specimen.J Med Life. 2022 Sep;15(9):1105-1109. doi: 10.25122/jml-2021-0196. J Med Life. 2022. PMID: 36415531 Free PMC article.
Cited by
-
Antibiotic Resistance Profile of Enterovirulent E. coli Isolates Harboring Broad-Spectrum Beta-Lactamase Genes in Cancer Patients at the Laquintinie Hospital in Douala, Littoral Region, Cameroon.Int J Microbiol. 2025 Jan 9;2025:4224807. doi: 10.1155/ijm/4224807. eCollection 2025. Int J Microbiol. 2025. PMID: 39822382 Free PMC article.
References
-
- Fongang H., Mbaveng A. T., Kuete V. Chapter one-global burden of bacterial infections and drug resistance. Advances in Botanical Research . 2023;106:1–20. doi: 10.1016/bs.abr.2022.08.001. - DOI
-
- Bonten M. J., Bergmans D. C., Speijer H., Stobberingh E. E. Characteristics of polyclonal endemicity of Pseudomonas aeruginosa colonization in intensive care units. Implications for infection control. American Journal of Respiratory and Critical Care Medicine . 1999;160(4):1212–1219. doi: 10.1164/ajrccm.160.4.9809031. - DOI - PubMed
LinkOut - more resources
Full Text Sources

