An Immunoinformatic-Based In Silico Identification on the Creation of a Multiepitope-Based Vaccination Against the Nipah Virus
- PMID: 38962403
- PMCID: PMC11221950
- DOI: 10.1155/2024/4066641
An Immunoinformatic-Based In Silico Identification on the Creation of a Multiepitope-Based Vaccination Against the Nipah Virus
Abstract
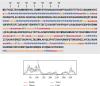
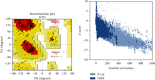
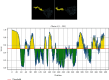
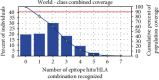
The zoonotic viruses pose significant threats to public health. Nipah virus (NiV) is an emerging virus transmitted from bats to humans. The NiV causes severe encephalitis and acute respiratory distress syndrome, leading to high mortality rates, with fatality rates ranging from 40% to 75%. The first emergence of the disease was found in Malaysia in 1998-1999 and later in Bangladesh, Cambodia, Timor-Leste, Indonesia, Singapore, Papua New Guinea, Vietnam, Thailand, India, and other South and Southeast Asian nations. Currently, no specific vaccines or antiviral drugs are available. The potential advantages of epitope-based vaccines include their ability to elicit specific immune responses while minimizing potential side effects. The epitopes have been identified from the conserved region of viral proteins obtained from the UniProt database. The selection of conserved epitopes involves analyzing the genetic sequences of various viral strains. The present study identified two B cell epitopes, seven cytotoxic T lymphocyte (CTL) epitopes, and seven helper T lymphocyte (HTL) epitope interactions from the NiV proteomic inventory. The antigenic and physiological properties of retrieved protein were analyzed using online servers ToxinPred, VaxiJen v2.0, and AllerTOP. The final vaccine candidate has a total combined coverage range of 80.53%. The tertiary structure of the constructed vaccine was optimized, and its stability was confirmed with the help of molecular simulation. Molecular docking was performed to check the binding affinity and binding energy of the constructed vaccine with TLR-3 and TLR-5. Codon optimization was performed in the constructed vaccine within the Escherichia coli K12 strain, to eliminate the danger of codon bias. However, these findings must require further validation to assess their effectiveness and safety. The development of vaccines and therapeutic approaches for virus infection is an ongoing area of research, and it may take time before effective interventions are available for clinical use.
Keywords: B cell epitope; Nipah virus; T cell epitope; epidemiology; epitope-based vaccine.
Copyright © 2024 Beant Kaur et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Developing a chimeric multiepitope vaccine against Nipah virus (NiV) through immunoinformatics, molecular docking and dynamic simulation approaches.Microb Pathog. 2024 Dec;197:107098. doi: 10.1016/j.micpath.2024.107098. Epub 2024 Nov 8. Microb Pathog. 2024. PMID: 39521154
-
Advancing one health vaccination: In silico design and evaluation of a multi-epitope subunit vaccine against Nipah virus for cross-species immunization using immunoinformatics and molecular modeling.PLoS One. 2024 Sep 26;19(9):e0310703. doi: 10.1371/journal.pone.0310703. eCollection 2024. PLoS One. 2024. PMID: 39325755 Free PMC article.
-
Comprehensive immunoinformatics and bioinformatics strategies for designing a multi-epitope based vaccine targeting structural proteins of Nipah virus.Front Immunol. 2025 May 13;16:1535322. doi: 10.3389/fimmu.2025.1535322. eCollection 2025. Front Immunol. 2025. PMID: 40433372 Free PMC article.
-
Nipah virus: epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies - a comprehensive review.Vet Q. 2019 Dec;39(1):26-55. doi: 10.1080/01652176.2019.1580827. Vet Q. 2019. PMID: 31006350 Free PMC article. Review.
-
Advances in diagnostics, vaccines and therapeutics for Nipah virus.Microbes Infect. 2019 Aug-Sep;21(7):278-286. doi: 10.1016/j.micinf.2019.02.002. Epub 2019 Feb 26. Microbes Infect. 2019. PMID: 30817995 Review.
Cited by
-
Repurposing of approved drugs towards Nipah virus treatment: an in silico docking, molecular dynamics simulation and a MM/GBSA approach.In Silico Pharmacol. 2025 Jun 13;13(2):86. doi: 10.1007/s40203-025-00371-z. eCollection 2025. In Silico Pharmacol. 2025. PMID: 40520959
-
Machine learning tools used for mapping some immunogenic epitopes within the major structural proteins of the bovine coronavirus (BCoV) and for the in silico design of the multiepitope-based vaccines.Front Vet Sci. 2024 Oct 2;11:1468890. doi: 10.3389/fvets.2024.1468890. eCollection 2024. Front Vet Sci. 2024. PMID: 39415947 Free PMC article.
-
Design and Preliminary Immunogenicity Evaluation of Nipah Virus Glycoprotein G Epitope-Based Peptide Vaccine in Mice.Vaccines (Basel). 2025 Apr 18;13(4):428. doi: 10.3390/vaccines13040428. Vaccines (Basel). 2025. PMID: 40333318 Free PMC article.
-
Computational design of multi-epitope vaccine against Hepatitis C Virus infection using immunoinformatics techniques.PLoS One. 2025 Jan 24;20(1):e0317520. doi: 10.1371/journal.pone.0317520. eCollection 2025. PLoS One. 2025. PMID: 39854342 Free PMC article.
References
-
- Sachan A., Singh R., Gupta B., Kulesh R. Nipah Virus and Its Zoonotic Importance: A Review. Journal of Entomology and Zoology Studies . 2023;11(1):208–213. doi: 10.22271/j.ento.2023.v11.i1c.9158. - DOI
-
- Luby S. P., Broder C. C. Viral Infections of Humans: Epidemiology and Control . New York, NY: Springer US; 2023. Paramyxoviruses: henipaviruses; pp. 1–51. - DOI
-
- Brown B., Gravier T., Fricke I., et al. Immunopathogenesis of Nipah virus infection and associated immune responses. Immuno . 2023;3(2):160–181. doi: 10.3390/immuno3020011. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

