Identification and immunological characterization of genes associated with ferroptosis in Alzheimer's disease and experimental demonstration
- PMID: 38963039
- PMCID: PMC11240865
- DOI: 10.3892/mmr.2024.13279
Identification and immunological characterization of genes associated with ferroptosis in Alzheimer's disease and experimental demonstration
Abstract
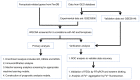
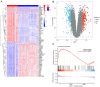
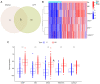
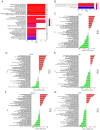
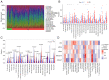
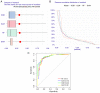
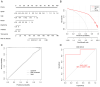
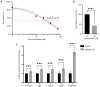
The incidence of Alzheimer's disease (AD) is rising globally, yet its treatment and prediction of this condition remain challenging due to the complex pathophysiological mechanisms associated with it. Consequently, the objective of the present study was to analyze and characterize the molecular mechanisms underlying ferroptosis‑related genes (FEGs) in the pathogenesis of AD, as well as to construct a prognostic model. The findings will provide new insights for the future diagnosis and treatment of AD. First, the AD dataset GSE33000 from the Gene Expression Omnibus database and the FEGs from FerrDB were obtained. Next, unsupervised cluster analysis was used to obtain the FEGs that were most relevant to AD. Subsequently, enrichment analyses were performed on the FEGs to explore biological functions. Subsequently, the role of these genes in the immune microenvironment was elucidated through CIBERSORT. Then, the optimal machine learning was selected by comparing the performance of different machine learning models. To validate the prediction efficiency, the models were validated using nomograms, calibration curves, decision curve analysis and external datasets. Furthermore, the expression of FEGs between different groups was verified using reverse transcription quantitative PCR and western blot analysis. In AD, alterations in the expression of FEGs affect the aggregation and infiltration of certain immune cells. This indicated that the occurrence of AD is strongly associated with immune infiltration. Finally, the most appropriate machine learning models were selected, and AD diagnostic models and nomograms were built. The present study provided novel insights that enhance understanding with regard to the molecular mechanism of action of FEGs in AD. Moreover, the present study provided biomarkers that may facilitate the diagnosis of AD.
Keywords: Alzheimer's disease; ferroptosis; immune infiltration; machine learning model; nomogram.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Deciphering the role of lipid metabolism-related genes in Alzheimer's disease: a machine learning approach integrating Traditional Chinese Medicine.Front Endocrinol (Lausanne). 2024 Oct 23;15:1448119. doi: 10.3389/fendo.2024.1448119. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39507054 Free PMC article.
-
Bioinformatics insights into mitochondrial and immune gene regulation in Alzheimer's disease.Eur J Med Res. 2025 Feb 8;30(1):89. doi: 10.1186/s40001-025-02297-w. Eur J Med Res. 2025. PMID: 39920860 Free PMC article.
-
Identifying the Role of Oligodendrocyte Genes in the Diagnosis of Alzheimer's Disease through Machine Learning and Bioinformatics Analysis.Curr Alzheimer Res. 2024;21(6):437-455. doi: 10.2174/0115672050338777241028071955. Curr Alzheimer Res. 2024. PMID: 39506420
-
Discovery and validation of Ferroptosis-related molecular patterns and immune characteristics in Alzheimer's disease.Front Aging Neurosci. 2022 Nov 23;14:1056312. doi: 10.3389/fnagi.2022.1056312. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36506471 Free PMC article.
-
Bioinformatics analysis and prediction of Alzheimer's disease and alcohol dependence based on Ferroptosis-related genes.Front Aging Neurosci. 2023 Jul 13;15:1201142. doi: 10.3389/fnagi.2023.1201142. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37520121 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

