A cargo sorting receptor mediates chloroplast protein trafficking through the secretory pathway
- PMID: 38963880
- PMCID: PMC11371137
- DOI: 10.1093/plcell/koae197
A cargo sorting receptor mediates chloroplast protein trafficking through the secretory pathway
Abstract
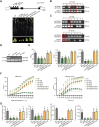
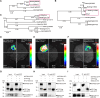
Nucleus-encoded chloroplast proteins can be transported via the secretory pathway. The molecular mechanisms underlying the trafficking of chloroplast proteins between the intracellular compartments are largely unclear, and a cargo sorting receptor has not previously been identified in the secretory pathway. Here, we report a cargo sorting receptor that is specifically present in Viridiplantae and mediates the transport of cargo proteins to the chloroplast. Using a forward genetic analysis, we identified a gene encoding a transmembrane protein (MtTP930) in barrel medic (Medicago truncatula). Mutation of MtTP930 resulted in impaired chloroplast function and a dwarf phenotype. MtTP930 is highly expressed in the aerial parts of the plant and is localized to the endoplasmic reticulum (ER) exit sites and Golgi. MtTP930 contains typical cargo sorting receptor motifs, interacts with Sar1, Sec12, and Sec24, and participates in coat protein complex II vesicular transport. Importantly, MtTP930 can recognize the cargo proteins plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase (MtNPP) and α-carbonic anhydrase (MtCAH) in the ER and then transport them to the chloroplast via the secretory pathway. Mutation of a homolog of MtTP930 in Arabidopsis (Arabidopsis thaliana) resulted in a similar dwarf phenotype. Furthermore, MtNPP-GFP failed to localize to chloroplasts when transgenically expressed in Attp930 protoplasts, implying that these cargo sorting receptors are conserved in plants. These findings fill a gap in our understanding of the mechanism by which chloroplast proteins are sorted and transported via the secretory pathway.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures






References
MeSH terms
Substances
Grants and funding
- 2023YFF1001400/National Key Research and Development Program of China
- 32001391/Young Scientists Fund of the National Natural Science Foundation of China
- BX20190371/China Postdoctoral Innovative Talents Support Program
- 2019M660876/China Postdoctoral Science Foundation
- 2022-101/College of Life Sciences, Inner Mongolia University
LinkOut - more resources
Full Text Sources
Miscellaneous

