SARS-CoV-2 genomic surveillance of migrants arriving to Europe through the Mediterranean routes
- PMID: 38963881
- PMCID: PMC11223754
- DOI: 10.7189/jogh.14.05017
SARS-CoV-2 genomic surveillance of migrants arriving to Europe through the Mediterranean routes
Abstract
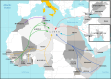
Background: The implementation genomic-based surveillance on emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants in low-income countries, which have inadequate molecular and sequencing capabilities and limited vaccine storage, represents a challenge for public health. To date, there is little evidence on molecular investigations of SARS-CoV-2 variants in areas where they might emerge. We report the findings of an experimental SARS-CoV-2 molecular surveillance programme for migrants, refugees, and asylum seekers arriving to Europe via Italy through the Mediterranean Sea.
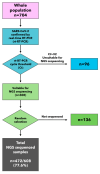
Methods: We descriptively analysed data on migrants collected at entry points in Sicily from February 2021 to May 2022. These entry points are integrated with a network of laboratories fully equipped for molecular analyses, which performed next-generation sequencing and used Nextclade and the Pangolin coronavirus disease 2019 (COVID-19) tools for clade/lineage assignment.
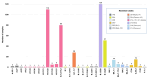
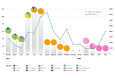
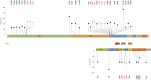
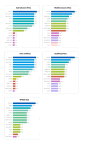
Results: We obtained 472 full-length SARS-CoV-2 sequences and identified 12 unique clades belonging to 31 different lineages. The delta variant accounted for 43.6% of all genomes, followed by clades 21D (Eta) and 20A (25.4% and 11.4%, respectively). Notably, some of the identified lineages (A.23.1, A.27, and A.29) predicted their introduction into the migration area. The mutation analysis allowed us to identify 617 different amino acid substitutions, 156 amino acid deletions, 7 stop codons, and 6 amino acid insertions. Lastly, we highlighted the geographical distribution patterns of some mutational profiles occurring in the migrants' countries of origin.
Conclusions: Genome-based molecular surveillance dedicated to migrant populations from low-resource areas may be useful for forecasting new epidemiological scenarios related to SARS-CoV-2 variants or other emerging pathogens, as well as for informing the updating of vaccination strategies.
Copyright © 2024 by the Journal of Global Health. All rights reserved.
Conflict of interest statement
Disclosure of interest: The authors completed the ICMJE Disclosure of Interest Form (available upon request from the corresponding author) and disclose no relevant interests.
Figures

References
-
- Italian Republic Ministry of Internal Affairs. Department of civilian freedom and migration. 2024. Available: http://www.libertaciviliimmigrazione.dlci.interno.gov.it/it/documentazio.... Accessed: 25 March 2024.
-
- International Organization for Migration (IOM). Report on the Central Mediterranean Route: Trends and Dynamics. 2015. Available: https://publications.iom.int/system/files/pdf/migration_data_on_the_cent.... Accessed: 25 March 2024.
-
- United Nations High Commissioner for Refugees (UNHCR). Desperate Journeys: Refugees and migrants arriving in Europe and at Europe's borders. 2018. Available: https://www.unhcr.org/desperatejourneys. Accessed: 25 March 2024.
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
