A data science roadmap for open science organizations engaged in early-stage drug discovery
- PMID: 38965235
- PMCID: PMC11224410
- DOI: 10.1038/s41467-024-49777-x
A data science roadmap for open science organizations engaged in early-stage drug discovery
Abstract
The Structural Genomics Consortium is an international open science research organization with a focus on accelerating early-stage drug discovery, namely hit discovery and optimization. We, as many others, believe that artificial intelligence (AI) is poised to be a main accelerator in the field. The question is then how to best benefit from recent advances in AI and how to generate, format and disseminate data to enable future breakthroughs in AI-guided drug discovery. We present here the recommendations of a working group composed of experts from both the public and private sectors. Robust data management requires precise ontologies and standardized vocabulary while a centralized database architecture across laboratories facilitates data integration into high-value datasets. Lab automation and opening electronic lab notebooks to data mining push the boundaries of data sharing and data modeling. Important considerations for building robust machine-learning models include transparent and reproducible data processing, choosing the most relevant data representation, defining the right training and test sets, and estimating prediction uncertainty. Beyond data-sharing, cloud-based computing can be harnessed to build and disseminate machine-learning models. Important vectors of acceleration for hit and chemical probe discovery will be (1) the real-time integration of experimental data generation and modeling workflows within design-make-test-analyze (DMTA) cycles openly, and at scale and (2) the adoption of a mindset where data scientists and experimentalists work as a unified team, and where data science is incorporated into the experimental design.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
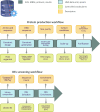
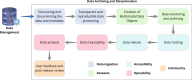

References
-
- For chemists, the AI revolution has yet to happen. Nature617, 438 (2023). - PubMed
-
- Guarino, N. Formal Ontology and Information Systems. (IOS Press 1998).
Publication types
MeSH terms
Grants and funding
- R01 GM140154/GM/NIGMS NIH HHS/United States
- T32 GM135122/GM/NIGMS NIH HHS/United States
- RGPIN-2019-04416/Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada (NSERC Canadian Network for Research and Innovation in Machining Technology)
LinkOut - more resources
Full Text Sources
Miscellaneous

