TWEAK/Fn14 signalling driven super-enhancer reprogramming promotes pro-metastatic metabolic rewiring in triple-negative breast cancer
- PMID: 38965263
- PMCID: PMC11224303
- DOI: 10.1038/s41467-024-50071-z
TWEAK/Fn14 signalling driven super-enhancer reprogramming promotes pro-metastatic metabolic rewiring in triple-negative breast cancer
Abstract
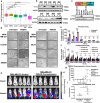
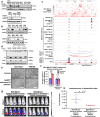
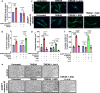
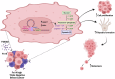
Triple Negative Breast Cancer (TNBC) is the most aggressive breast cancer subtype suffering from limited targeted treatment options. Following recent reports correlating Fibroblast growth factor-inducible 14 (Fn14) receptor overexpression in Estrogen Receptor (ER)-negative breast cancers with metastatic events, we show that Fn14 is specifically overexpressed in TNBC patients and associated with poor survival. We demonstrate that constitutive Fn14 signalling rewires the transcriptomic and epigenomic landscape of TNBC, leading to enhanced tumour growth and metastasis. We further illustrate that such mechanisms activate TNBC-specific super enhancers (SE) to drive the transcriptional activation of cancer dependency genes via chromatin looping. In particular, we uncover the SE-driven upregulation of Nicotinamide phosphoribosyltransferase (NAMPT), which promotes NAD+ and ATP metabolic reprogramming critical for filopodia formation and metastasis. Collectively, our study details the complex mechanistic link between TWEAK/Fn14 signalling and TNBC metastasis, which reveals several vulnerabilities which could be pursued for the targeted treatment of TNBC patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

