A CRISPR activation screen identifies FBXO22 supporting targeted protein degradation
- PMID: 38965383
- PMCID: PMC11581908
- DOI: 10.1038/s41589-024-01655-9
A CRISPR activation screen identifies FBXO22 supporting targeted protein degradation
Abstract
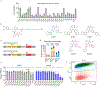
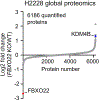
Targeted protein degradation (TPD) represents a potent chemical biology paradigm that leverages the cellular degradation machinery to pharmacologically eliminate specific proteins of interest. Although multiple E3 ligases have been discovered to facilitate TPD, there exists a compelling requirement to diversify the pool of E3 ligases available for such applications. Here we describe a clustered regularly interspaced short palindromic repeats (CRISPR)-based transcriptional activation screen focused on human E3 ligases, with the goal of identifying E3 ligases that can facilitate heterobifunctional compound-mediated target degradation. Through this approach, we identified a candidate proteolysis-targeting chimera (PROTAC), 22-SLF, that induces the degradation of FK506-binding protein 12 when the transcription of FBXO22 gene is activated. Subsequent mechanistic investigations revealed that 22-SLF interacts with C227 and/or C228 in F-box protein 22 (FBXO22) to achieve target degradation. Lastly, we demonstrated the versatility of FBXO22-based PROTACs by effectively degrading additional endogenous proteins, including bromodomain-containing protein 4 and the echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase fusion protein.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: A.A.B. and X.Z. are named on a patent application related to TPD, held by Northwestern University (US provisional patent application number 63/538,637). The other authors declare no competing interests.
Figures


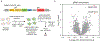



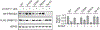




Update of
-
A CRISPR activation screen identifies FBXO22 as an E3 ligase supporting targeted protein degradation.bioRxiv [Preprint]. 2023 Sep 16:2023.09.15.557708. doi: 10.1101/2023.09.15.557708. bioRxiv. 2023. Update in: Nat Chem Biol. 2024 Dec;20(12):1608-1616. doi: 10.1038/s41589-024-01655-9. PMID: 37745578 Free PMC article. Updated. Preprint.
References
-
- Kannt A. & Dikic I. Expanding the arsenal of E3 ubiquitin ligases for proximity-induced protein degradation. Cell Chem Biol 28, 1014–1031 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- T32 GM105538/GM/NIGMS NIH HHS/United States
- R00 CA248715/CA/NCI NIH HHS/United States
- T32GM149439/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R00CA248715/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- DFS-53-22/Damon Runyon Cancer Research Foundation (Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation)
LinkOut - more resources
Full Text Sources

