An novel effective and safe model for the diagnosis of nonalcoholic fatty liver disease in China: gene excavations, clinical validations, and mechanism elucidation
- PMID: 38965537
- PMCID: PMC11225259
- DOI: 10.1186/s12967-024-05315-3
An novel effective and safe model for the diagnosis of nonalcoholic fatty liver disease in China: gene excavations, clinical validations, and mechanism elucidation
Abstract
Background: Non-alcoholic fatty liver disease (NAFLD) is one of the most common chronic liver diseases. NAFLD leads to liver fibrosis and hepatocellular carcinoma, and it also has systemic effects associated with metabolic diseases, cardiovascular diseases, chronic kidney disease, and malignant tumors. Therefore, it is important to diagnose NAFLD early to prevent these adverse effects.
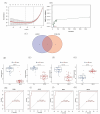
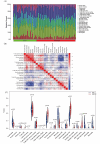
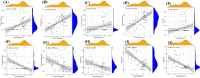
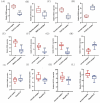
Methods: The GSE89632 dataset was downloaded from the Gene Expression Omnibus database, and then the optimal genes were screened from the data cohort using lasso and Support Vector Machine Recursive Feature Elimination (SVM-RFE). The ROC values of the optimal genes for the diagnosis of NAFLD were calculated. The relationship between optimal genes and immune cells was determined using the DECONVOLUTION algorithm CIBERSORT. Finally, the specificity and sensitivity of the diagnostic genes were verified by detecting the expression of the diagnostic genes in blood samples from 320 NAFLD patients and liver samples from 12 mice.
Results: Through machine learning we identified FOSB, GPAT3, RGCC and RNF43 were the key diagnostic genes for NAFLD, and they were further demonstrated by a receiver operating characteristic curve analysis. We found that the combined diagnosis of the four genes identified NAFLD samples well from normal samples (AUC = 0.997). FOSB, GPAT3, RGCC and RNF43 were strongly associated with immune cell infiltration. We also experimentally examined the expression of these genes in NAFLD patients and NAFLD mice, and the results showed that these genes are highly specific and sensitive.
Conclusions: Data from both clinical and animal studies demonstrate the high sensitivity, specificity and safety of FOSB, GPAT3, RGCC and RNF43 for the diagnosis of NAFLD. The relationship between diagnostic key genes and immune cell infiltration may help to understand the development of NAFLD. The study was reviewed and approved by Ethics Committee of Tianjin Second People's Hospital in 2021 (ChiCTR1900024415).
Keywords: Biomarkers; Machine learning; Nonalcoholic fatty liver disease.
© 2024. The Author(s).
Conflict of interest statement
No conflict of interest exits in the submission of this manuscript, and manuscript is approved by all authors for publication.
Authors declare that they have no competing interests.
Figures





Similar articles
-
Identification of common signature genes and pathways underlying the pathogenesis association between nonalcoholic fatty liver disease and heart failure.Front Immunol. 2024 Sep 16;15:1424308. doi: 10.3389/fimmu.2024.1424308. eCollection 2024. Front Immunol. 2024. PMID: 39351239 Free PMC article.
-
Integrative analysis identifies oxidative stress biomarkers in non-alcoholic fatty liver disease via machine learning and weighted gene co-expression network analysis.Front Immunol. 2024 Feb 27;15:1335112. doi: 10.3389/fimmu.2024.1335112. eCollection 2024. Front Immunol. 2024. PMID: 38476236 Free PMC article.
-
Identification of potential metabolic biomarkers and immune cell infiltration for metabolic associated steatohepatitis by bioinformatics analysis and machine learning.Sci Rep. 2025 May 13;15(1):16596. doi: 10.1038/s41598-025-86397-x. Sci Rep. 2025. PMID: 40360670 Free PMC article.
-
Identification of potential feature genes in non-alcoholic fatty liver disease using bioinformatics analysis and machine learning strategies.Comput Biol Med. 2023 May;157:106724. doi: 10.1016/j.compbiomed.2023.106724. Epub 2023 Mar 5. Comput Biol Med. 2023. PMID: 36898287
-
Identification and validation of potential diagnostic signature and immune cell infiltration for NAFLD based on cuproptosis-related genes by bioinformatics analysis and machine learning.Front Immunol. 2023 Sep 26;14:1251750. doi: 10.3389/fimmu.2023.1251750. eCollection 2023. Front Immunol. 2023. PMID: 37822923 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

