GNA13 suppresses proliferation of ER+ breast cancer cells via ERα dependent upregulation of the MYC oncogene
- PMID: 38965558
- PMCID: PMC11225210
- DOI: 10.1186/s13058-024-01866-x
GNA13 suppresses proliferation of ER+ breast cancer cells via ERα dependent upregulation of the MYC oncogene
Abstract
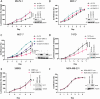
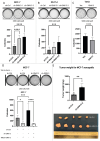
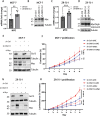
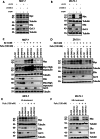
GNA13 (Gα13) is one of two alpha subunit members of the G12/13 family of heterotrimeric G-proteins which mediate signaling downstream of GPCRs. It is known to be essential for embryonic development and vasculogenesis and has been increasingly shown to be involved in mediating several steps of cancer progression. Recent studies found that Gα13 can function as an oncogene and contributes to progression and metastasis of multiple tumor types, including ovarian, head and neck and prostate cancers. In most cases, Gα12 and Gα13, as closely related α-subunits in the subfamily, have similar cellular roles. However, in recent years their differences in signaling and function have started to emerge. We previously identified that Gα13 drives invasion of Triple Negative Breast Cancer (TNBC) cells in vitro. As a highly heterogenous disease with various well-defined molecular subtypes (ER+ /Her2-, ER+ /Her2+, Her2+, TNBC) and subtype associated outcomes, the function(s) of Gα13 beyond TNBC should be explored. Here, we report the finding that low expression of GNA13 is predictive of poorer survival in breast cancer, which challenges the conventional idea of Gα12/13 being universal oncogenes in solid tumors. Consistently, we found that Gα13 suppresses the proliferation in multiple ER+ breast cancer cell lines (MCF-7, ZR-75-1 and T47D). Loss of GNA13 expression drives cell proliferation, soft-agar colony formation and in vivo tumor formation in an orthotopic xenograft model. To evaluate the mechanism of Gα13 action, we performed RNA-sequencing analysis on these cell lines and found that loss of GNA13 results in the upregulation of MYC signaling pathways in ER+ breast cancer cells. Simultaneous silencing of MYC reversed the proliferative effect from the loss of GNA13, validating the role of MYC in Gα13 regulation of proliferation. Further, we found Gα13 regulates the expression of MYC, at both the transcript and protein level in an ERα dependent manner. Taken together, our study provides the first evidence for a tumor suppressive role for Gα13 in breast cancer cells and demonstrates for the first time the direct involvement of Gα13 in ER-dependent regulation of MYC signaling. With a few exceptions, elevated Gα13 levels are generally considered to be oncogenic, similar to Gα12. This study demonstrates an unexpected tumor suppressive role for Gα13 in ER+ breast cancer via regulation of MYC, suggesting that Gα13 can have subtype-dependent tumor suppressive roles in breast cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The GNA13-RhoA signaling axis suppresses expression of tumor protective Kallikreins.Cell Signal. 2016 Oct;28(10):1479-88. doi: 10.1016/j.cellsig.2016.07.001. Epub 2016 Jul 14. Cell Signal. 2016. PMID: 27424208
-
Context-dependent roles of MDMX (MDM4) and MDM2 in breast cancer proliferation and circulating tumor cells.Breast Cancer Res. 2019 Jan 14;21(1):5. doi: 10.1186/s13058-018-1094-8. Breast Cancer Res. 2019. PMID: 30642351 Free PMC article.
-
Inactivating mutations in GNA13 and RHOA in Burkitt's lymphoma and diffuse large B-cell lymphoma: a tumor suppressor function for the Gα13/RhoA axis in B cells.Oncogene. 2016 Jul 21;35(29):3771-80. doi: 10.1038/onc.2015.442. Epub 2015 Nov 30. Oncogene. 2016. PMID: 26616858 Free PMC article.
-
GPCR-Gα13 Involvement in Mitochondrial Function, Oxidative Stress, and Prostate Cancer.Int J Mol Sci. 2024 Jun 28;25(13):7162. doi: 10.3390/ijms25137162. Int J Mol Sci. 2024. PMID: 39000269 Free PMC article. Review.
-
The emerging roles of Gα12/13 proteins on the hallmarks of cancer in solid tumors.Oncogene. 2022 Jan;41(2):147-158. doi: 10.1038/s41388-021-02069-w. Epub 2021 Oct 23. Oncogene. 2022. PMID: 34689178 Free PMC article. Review.
References
-
- Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. - DOI - PMC - PubMed
-
- Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9:176–198. doi: 10.1158/2159-8290.CD-18-1177. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

