A novel lncRNA LOC101928222 promotes colorectal cancer angiogenesis by stabilizing HMGCS2 mRNA and increasing cholesterol synthesis
- PMID: 38965575
- PMCID: PMC11223299
- DOI: 10.1186/s13046-024-03095-8
A novel lncRNA LOC101928222 promotes colorectal cancer angiogenesis by stabilizing HMGCS2 mRNA and increasing cholesterol synthesis
Abstract
Background: Metastasis is the leading cause of mortality in patients with colorectal cancer (CRC) and angiogenesis is a crucial factor in tumor invasion and metastasis. Long noncoding RNAs (lncRNAs) play regulatory functions in various biological processes in tumor cells, however, the roles of lncRNAs in CRC-associated angiogenesis remain to be elucidated in CRC, as do the underlying mechanisms.
Methods: We used bioinformatics to screen differentially expressed lncRNAs from TCGA database. LOC101928222 expression was assessed by qRT-PCR. The impact of LOC101928222 in CRC tumor development was assessed both in vitro and in vivo. The regulatory mechanisms of LOC101928222 in CRC were investigated by cellular fractionation, RNA-sequencing, mass spectrometric, RNA pull-down, RNA immunoprecipitation, RNA stability, and gene-specific m6A assays.
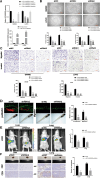
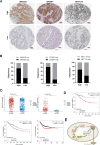
Results: LOC101928222 expression was upregulated in CRC and was correlated with a worse outcome. Moreover, LOC101928222 was shown to promote migration, invasion, and angiogenesis in CRC. Mechanistically, LOC101928222 synergized with IGF2BP1 to stabilize HMGCS2 mRNA through an m6A-dependent pathway, leading to increased cholesterol synthesis and, ultimately, the promotion of CRC development.
Conclusions: In summary, these findings demonstrate a novel, LOC101928222-based mechanism involved in the regulation of cholesterol synthesis and the metastatic potential of CRC. The LOC101928222-HMGCS2-cholesterol synthesis pathway may be an effective target for diagnosing and managing CRC metastasis.
Keywords: Angiogenesis; Cholesterol synthesis; Colorectal cancer; LOC101928222; m6A.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
LINC02167 stabilizes KSR1 mRNA in an m5C-dependent manner to regulate the ERK/MAPK signaling pathway and promotes colorectal cancer metastasis.J Exp Clin Cancer Res. 2025 Apr 15;44(1):121. doi: 10.1186/s13046-025-03368-w. J Exp Clin Cancer Res. 2025. PMID: 40234937 Free PMC article.
-
Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA.Mol Cancer. 2022 Aug 19;21(1):168. doi: 10.1186/s12943-022-01638-1. Mol Cancer. 2022. PMID: 35986274 Free PMC article.
-
LncRNA SNHG1 facilitates colorectal cancer cells metastasis by recruiting HNRNPD protein to stabilize SERPINA3 mRNA.Cancer Lett. 2024 Nov 1;604:217217. doi: 10.1016/j.canlet.2024.217217. Epub 2024 Sep 2. Cancer Lett. 2024. PMID: 39233042
-
Roles of long noncoding RNAs in colorectal cancer metastasis.Oncotarget. 2017 Jun 13;8(24):39859-39876. doi: 10.18632/oncotarget.16339. Oncotarget. 2017. PMID: 28418892 Free PMC article. Review.
-
LncRNAs: the bridge linking RNA and colorectal cancer.Oncotarget. 2017 Feb 14;8(7):12517-12532. doi: 10.18632/oncotarget.13573. Oncotarget. 2017. PMID: 27888635 Free PMC article. Review.
Cited by
-
Comprehensive analysis of cholesterol metabolism-related genes in prostate cancer: integrated analysis of single-cell and bulk RNA sequencing.Discov Oncol. 2025 Jul 30;16(1):1442. doi: 10.1007/s12672-025-03294-5. Discov Oncol. 2025. PMID: 40736630 Free PMC article.
-
METTL16 Promotes Lipid Metabolic Reprogramming and Colorectal Cancer Progression.Int J Biol Sci. 2025 Jul 24;21(11):4782-4797. doi: 10.7150/ijbs.105391. eCollection 2025. Int J Biol Sci. 2025. PMID: 40860196 Free PMC article.
-
Methylation-mediated LncRNA CRAT40 promotes colorectal cancer progression by recruiting YBX1 to initiate RelA transcription.Int J Biol Sci. 2025 Jul 25;21(11):4834-4850. doi: 10.7150/ijbs.105629. eCollection 2025. Int J Biol Sci. 2025. PMID: 40860198 Free PMC article.
-
LINC02167 stabilizes KSR1 mRNA in an m5C-dependent manner to regulate the ERK/MAPK signaling pathway and promotes colorectal cancer metastasis.J Exp Clin Cancer Res. 2025 Apr 15;44(1):121. doi: 10.1186/s13046-025-03368-w. J Exp Clin Cancer Res. 2025. PMID: 40234937 Free PMC article.
-
LncRNAs in oncogenic microenvironment: from threat to therapy.Front Cell Dev Biol. 2025 Mar 13;12:1423279. doi: 10.3389/fcell.2024.1423279. eCollection 2024. Front Cell Dev Biol. 2025. PMID: 40176927 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 81972278/National Natural Science Foundation of China
- 789ZYRC202090147/789 Outstanding Talent Program of SAHNMU
- LGY2019078/"Six One Project" Top Talent Program for High-level Health Talents in 2019
- JSDW202235/Jiangsu Province Capability Improvement Project through Science, Technology and Education; Jiangsu Provincial Medical Key Discipline Cultivation Unit
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

