Promoter profiles in plasma CfDNA exhibits a potential utility of predicting the efficacy of neoadjuvant chemotherapy in breast cancer patients
- PMID: 38965610
- PMCID: PMC11225256
- DOI: 10.1186/s13058-024-01860-3
Promoter profiles in plasma CfDNA exhibits a potential utility of predicting the efficacy of neoadjuvant chemotherapy in breast cancer patients
Abstract
Background: Gene expression profiles in breast tissue biopsies contain information related to chemotherapy efficacy. The promoter profiles in cell-free DNA (cfDNA) carrying gene expression information of the original tissues may be used to predict the response to neoadjuvant chemotherapy in breast cancer as a non-invasive biomarker. In this study, the feasibility of the promoter profiles in plasma cfDNA was evaluated as a novel clinical model for noninvasively predicting the efficacy of neoadjuvant chemotherapy in breast cancer.
Method: First of all, global chromatin (5 Mb windows), sub-compartments and promoter profiles in plasma cfDNA samples from 94 patients with breast cancer before neoadjuvant chemotherapy (pCR = 31 vs. non-pCR = 63) were analyzed, and then classifiers were developed for predicting the efficacy of neoadjuvant chemotherapy in breast cancer. Further, the promoter profile changes in sequential cfDNA samples from 30 patients (pCR = 8 vs. non-pCR = 22) during neoadjuvant chemotherapy were analyzed to explore the potential benefits of cfDNA promoter profile changes as a novel potential biomarker for predicting the treatment efficacy.
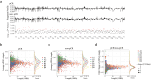
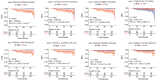
Results: The results showed significantly distinct promoter profile in plasma cfDNA of pCR patients compared with non-pCR patients before neoadjuvant chemotherapy. The classifier based on promoter profiles in a Random Forest model produced the largest area under the curve of 0.980 (95% CI: 0.978-0.983). After neoadjuvant chemotherapy, 332 genes with significantly differential promoter profile changes in sequential cfDNA samples of pCR patients was observed, compared with non-pCR patients, and their functions were closely related to treatment response.
Conclusion: These results suggest that promoter profiles in plasma cfDNA may be a powerful, non-invasive tool for predicting the efficacy of neoadjuvant chemotherapy breast cancer patients before treatment, and the on-treatment cfDNA promoter profiles have potential benefits for predicting the treatment efficacy.
Keywords: Neoadjuvant chemotherapy; Non-invasive; Plasma cfDNA; Promoter profiles; Treatment efficacy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Identification of the plasma total cfDNA level before and after chemotherapy as an indicator of the neoadjuvant chemotherapy response in locally advanced breast cancer.Cancer Med. 2020 Apr;9(7):2271-2282. doi: 10.1002/cam4.2906. Epub 2020 Feb 3. Cancer Med. 2020. PMID: 32017472 Free PMC article.
-
Measuring cfDNA integrity as a biomarker for predicting neoadjuvant chemotherapy response in breast cancer patients: a pilot study.Breast Cancer Res Treat. 2024 Jul;206(2):329-335. doi: 10.1007/s10549-024-07366-y. Epub 2024 May 14. Breast Cancer Res Treat. 2024. PMID: 38743176
-
Plasma Cell-Free DNA Concentration and Fragmentomes Predict Neoadjuvant Chemotherapy Response in Cervical Cancer Patients.Adv Sci (Weinh). 2024 Nov;11(43):e2309422. doi: 10.1002/advs.202309422. Epub 2024 Sep 25. Adv Sci (Weinh). 2024. PMID: 39319610 Free PMC article.
-
Serum Tumor Marker Levels might have Little Significance in Evaluating Neoadjuvant Treatment Response in Locally Advanced Breast Cancer.Asian Pac J Cancer Prev. 2015;16(11):4603-8. doi: 10.7314/apjcp.2015.16.11.4603. Asian Pac J Cancer Prev. 2015. PMID: 26107211
-
Predictive analysis of breast cancer response to neoadjuvant chemotherapy through plasma metabolomics.Breast Cancer Res Treat. 2024 Sep;207(2):393-404. doi: 10.1007/s10549-024-07370-2. Epub 2024 May 13. Breast Cancer Res Treat. 2024. PMID: 38740665
References
-
- Kuerer HM, Newman LA, Smith TL, Ames FC, Hunt KK, Dhingra K, Theriault RL, Singh G, Binkley SM, Sneige N, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17(2):460–9. doi: 10.1200/JCO.1999.17.2.460. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

