A colloidal model for the equilibrium assembly and liquid-liquid phase separation of the reflectin A1 protein
- PMID: 38965780
- PMCID: PMC11427776
- DOI: 10.1016/j.bpj.2024.07.004
A colloidal model for the equilibrium assembly and liquid-liquid phase separation of the reflectin A1 protein
Abstract
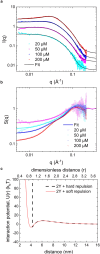
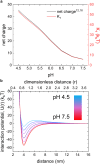
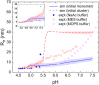
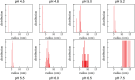
Reflectin is an intrinsically disordered protein known for its ability to modulate the biophotonic camouflage of cephalopods based on its assembly-induced osmotic properties. Its reversible self-assembly into discrete, size-controlled clusters and condensed droplets are known to depend sensitively on the net protein charge, making reflectin stimuli-responsive to pH, phosphorylation, and electric fields. Despite considerable efforts to characterize this behavior, the detailed physical mechanisms of reflectin's assembly are not yet fully understood. Here, we pursue a coarse-grained molecular understanding of reflectin assembly using a combination of experiments and simulations. We hypothesize that reflectin assembly and phase behavior can be explained from a remarkably simple colloidal model whereby individual protein monomers effectively interact via a short-range attractive and long-range repulsive (SA-LR) pair potential. We parameterize a coarse-grained SA-LR interaction potential for reflectin A1 from small-angle x-ray scattering measurements, and then extend it to a range of pH values using Gouy-Chapman theory to model monomer-monomer electrostatic interactions. The pH-dependent SA-LR interaction is then used in molecular dynamics simulations of reflectin assembly, which successfully capture a number of qualitative features of reflectin, including pH-dependent formation of discrete-sized nanoclusters and liquid-liquid phase separation at high pH, resulting in a putative phase diagram for reflectin. Importantly, we find that at low pH size-controlled reflectin clusters are equilibrium assemblies, which dynamically exchange protein monomers to maintain an equilibrium size distribution. These findings provide a mechanistic understanding of the equilibrium assembly of reflectin, and suggest that colloidal-scale models capture key driving forces and interactions to explain thermodynamic aspects of native reflectin behavior. Furthermore, the success of SA-LR interactions presented in this study demonstrates the potential of a colloidal interpretation of interactions and phenomena in a range of intrinsically disordered proteins.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Brangwynne C.P., Tompa P., Pappu R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015;11:899–904.
-
- Weber S.C. Sequence-encoded material properties dictate the structure and function of nuclear bodies. Curr. Opin. Cell Biol. 2017;46:62–71. - PubMed
-
- Sreekumar A., Nyati M.K., et al. Chinnaiyan A.M. Profiling of cancer cells using protein microarrays: Discovery of novel radiation-regulated proteins. Cancer Res. 2001;61:7585–7593. - PubMed
-
- Song J., Levenson R., et al. Morse D.E. Reflectin Proteins Bind and Reorganize Synthetic Phospholipid Vesicles. Langmuir. 2020;36:2673–2682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

