The two-faced role of RNA methyltransferase METTL3 on cellular response to cisplatin in head and neck squamous cell carcinoma in vitro model
- PMID: 38966069
- PMCID: PMC11223524
- DOI: 10.3389/fonc.2024.1402126
The two-faced role of RNA methyltransferase METTL3 on cellular response to cisplatin in head and neck squamous cell carcinoma in vitro model
Abstract
Background: RNA methyltransferase-like 3 (METTL3) is responsible for methyl group transfer in the progression of N 6-methyladenosine (m6A) modification. This epigenetic feature contributes to the structural and functional regulation of RNA and consequently may promote tumorigenesis, tumor progression, and cellular response to anticancer treatment (chemo-, radio-, and immunotherapy). In head and neck squamous cell carcinoma (HNSCC), the commonly used chemotherapy is cisplatin. Unfortunately, cisplatin resistance is still a major cause of tumor relapse and patients' death. Thus, this study aimed to investigate the role of METTL3 on cellular response to cisplatin in HNSCC in vitro models.
Materials and methods: HNSCC cell lines (H103, FaDu, and Detroit-562) with stable METTL3 knockdown (sgMETTL3) established with CRISPR-Cas9 system were treated with 0.5 tolerable plasma level (TPL) and 1 TPL of cisplatin. Further, cell cycle distribution, apoptosis, CD44/CD133 surface marker expression, and cell's ability to colony formation were analyzed in comparison to controls (cells transduced with control sgRNA).
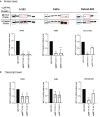
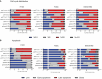
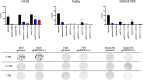
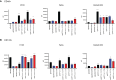
Results: The analyses of cell cycle distribution and apoptosis indicated a significantly higher percentage of cells with METTL3 knockdown 1) arrested in the G2/S phase and 2) characterized as a late apoptotic or death in comparison to control. The colony formation assay showed intensified inhibition of a single cell's ability to grow into a colony in FaDu and Detroit-562 METTL3-deficient cells, while a higher colony number was observed in H103 METTL3 knockdown cells after cisplatin treatment. Also, METTL3 deficiency significantly increased cancer stem cell markers' surface expression in all studied cell lines.
Conclusion: Our findings highlight the significant influence of METTL3 on the cellular response to cisplatin, suggesting its potential as a promising therapeutic target for addressing cisplatin resistance in certain cases of HNSCC.
Keywords: RNA methylation; RNA methyltransferase-like 3; chemotherapy; cisplatin; head and neck squamous cell carcinoma.
Copyright © 2024 Ostrowska, Rawłuszko-Wieczorek, Ostapowicz, Suchorska and Golusiński.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
Similar articles
-
METTL3 modulates m6A modification of CDC25B and promotes head and neck squamous cell carcinoma malignant progression.Exp Hematol Oncol. 2022 Mar 14;11(1):14. doi: 10.1186/s40164-022-00256-3. Exp Hematol Oncol. 2022. PMID: 35287752 Free PMC article.
-
Methyltransferase-like 3-induced N6-methyladenosine upregulation promotes oral squamous cell carcinoma by through p38.Oral Dis. 2023 Mar;29(2):639-648. doi: 10.1111/odi.14016. Epub 2021 Sep 12. Oral Dis. 2023. PMID: 34479400
-
METTL3-mediated ALDH m6A methylation regulates the malignant behavior of BMI1+ HNSCC stem cells.Oral Dis. 2024 Apr;30(3):1061-1071. doi: 10.1111/odi.14609. Epub 2023 May 30. Oral Dis. 2024. PMID: 37249063
-
C-Met pathway promotes self-renewal and tumorigenecity of head and neck squamous cell carcinoma stem-like cell.Oral Oncol. 2014 Jul;50(7):633-9. doi: 10.1016/j.oraloncology.2014.04.004. Epub 2014 May 15. Oral Oncol. 2014. PMID: 24835851 Review.
-
Novel insights into the m6A-RNA methyltransferase METTL3 in cancer.Biomark Res. 2021 Apr 20;9(1):27. doi: 10.1186/s40364-021-00278-9. Biomark Res. 2021. PMID: 33879256 Free PMC article. Review.
Cited by
-
Unveiling the impact of CD133 on cell cycle regulation in radio- and chemo-resistance of cancer stem cells.Front Public Health. 2025 Feb 6;13:1509675. doi: 10.3389/fpubh.2025.1509675. eCollection 2025. Front Public Health. 2025. PMID: 39980929 Free PMC article. Review.
-
Writers, readers, and erasers RNA modifications and drug resistance in cancer.Mol Cancer. 2024 Aug 30;23(1):178. doi: 10.1186/s12943-024-02089-6. Mol Cancer. 2024. PMID: 39215288 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

