Human CD56+CD39+ dNK cells support fetal survival through controlling trophoblastic cell fate: immune mechanisms of recurrent early pregnancy loss
- PMID: 38966071
- PMCID: PMC11223582
- DOI: 10.1093/nsr/nwae142
Human CD56+CD39+ dNK cells support fetal survival through controlling trophoblastic cell fate: immune mechanisms of recurrent early pregnancy loss
Abstract
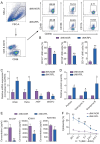
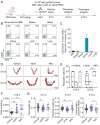
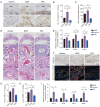
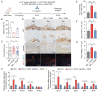
Decidual natural killer (dNK) cells are the most abundant immune cells at the maternal-fetal interface during early pregnancy in both mice and humans, and emerging single-cell transcriptomic studies have uncovered various human dNK subsets that are disrupted in patients experiencing recurrent early pregnancy loss (RPL) at early gestational stage, suggesting a connection between abnormal proportions or characteristics of dNK subsets and RPL pathogenesis. However, the functional mechanisms underlying this association remain unclear. Here, we established a mouse model by adoptively transferring human dNK cells into pregnant NOG (NOD/Shi-scid/IL-2Rγnull) mice, where human dNK cells predominantly homed into the uteri of recipients. Using this model, we observed a strong correlation between the properties of human dNK cells and pregnancy outcome. The transfer of dNK cells from RPL patients (dNK-RPL) remarkably worsened early pregnancy loss and impaired placental trophoblast cell differentiation in the recipients. These adverse effects were effectively reversed by transferring CD56+CD39+ dNK cells. Mechanistic studies revealed that CD56+CD39+ dNK subset facilitates early differentiation of mouse trophoblast stem cells (mTSCs) towards both invasive and syncytial pathways through secreting macrophage colony-stimulating factor (M-CSF). Administration of recombinant M-CSF to NOG mice transferred with dNK-RPL efficiently rescued the exacerbated pregnancy outcomes and fetal/placental development. Collectively, this study established a novel humanized mouse model featuring functional human dNK cells homing into the uteri of recipients and uncovered the pivotal role of M-CSF in fetal-supporting function of CD56+CD39+ dNK cells during early pregnancy, highlighting that M-CSF may be a previously unappreciated therapeutic target for intervening RPL.
Keywords: CD56+CD39+ dNK subset; M-CSF; NOG mice; adoptive transfer; placental trophoblast cell fate; recurrent pregnancy loss.
© The Author(s) 2024. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures

Similar articles
-
Decidual CXCR4+ CD56bright NK cells as a novel NK subset in maternal-foetal immune tolerance to alleviate early pregnancy failure.Clin Transl Med. 2021 Oct;11(10):e540. doi: 10.1002/ctm2.540. Clin Transl Med. 2021. PMID: 34709764 Free PMC article.
-
The immune potential of decidua-resident CD16+CD56+ NK cells in human pregnancy.Hum Immunol. 2021 May;82(5):332-339. doi: 10.1016/j.humimm.2021.01.014. Epub 2021 Feb 12. Hum Immunol. 2021. PMID: 33583640
-
CD56(bright)CD25+ NK cells are preferentially recruited to the maternal/fetal interface in early human pregnancy.Cell Mol Immunol. 2015 Jan;12(1):77-86. doi: 10.1038/cmi.2014.26. Epub 2014 May 5. Cell Mol Immunol. 2015. PMID: 24793405 Free PMC article.
-
Decidual Natural Killer Cells: A Good Nanny at the Maternal-Fetal Interface During Early Pregnancy.Front Immunol. 2021 May 12;12:663660. doi: 10.3389/fimmu.2021.663660. eCollection 2021. Front Immunol. 2021. PMID: 34054831 Free PMC article. Review.
-
Killer Timing: The Temporal Uterine Natural Killer Cell Differentiation Pathway and Implications for Female Reproductive Health.Front Endocrinol (Lausanne). 2022 Jun 27;13:904744. doi: 10.3389/fendo.2022.904744. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35832424 Free PMC article. Review.
Cited by
-
Hypoxia-Induced O-GlcNAcylation of GATA3 Leads to Excessive Testosterone Production in Preeclamptic Placentas.MedComm (2020). 2025 Feb 23;6(3):e70115. doi: 10.1002/mco2.70115. eCollection 2025 Mar. MedComm (2020). 2025. PMID: 39991625 Free PMC article.
-
Dysregulation of MYBL2 impairs extravillous trophoblast lineage development and function, contributing to recurrent spontaneous abortion.Proc Natl Acad Sci U S A. 2025 May 6;122(18):e2421653122. doi: 10.1073/pnas.2421653122. Epub 2025 Apr 28. Proc Natl Acad Sci U S A. 2025. PMID: 40294258
-
JEG-3 Trophoblast Cells Influence ILC-like Transformation of NK Cells In Vitro.Int J Mol Sci. 2025 Apr 14;26(8):3687. doi: 10.3390/ijms26083687. Int J Mol Sci. 2025. PMID: 40332223 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
