Simultaneous detection of 5-methylcytosine and 5-hydroxymethylcytosine at specific genomic loci by engineered deaminase-assisted sequencing
- PMID: 38966352
- PMCID: PMC11220598
- DOI: 10.1039/d4sc00930d
Simultaneous detection of 5-methylcytosine and 5-hydroxymethylcytosine at specific genomic loci by engineered deaminase-assisted sequencing
Abstract
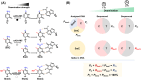
Cytosine modifications, particularly 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), play crucial roles in numerous biological processes. Current analytical methods are often constrained to the separate detection of either 5mC or 5hmC, or the combination of both modifications. The ability to simultaneously detect C, 5mC, and 5hmC at the same genomic locations with precise stoichiometry is highly desirable. Herein, we introduce a method termed engineered deaminase-assisted sequencing (EDA-seq) for the simultaneous quantification of C, 5mC, and 5hmC at the same genomic sites. EDA-seq utilizes a specially engineered protein, derived from human APOBEC3A (A3A), known as eA3A-M5. eA3A-M5 exhibits distinct deamination capabilities for C, 5mC, and 5hmC. In EDA-seq, C undergoes complete deamination and is sequenced as T. 5mC is partially deaminated resulting in a mixed readout of T and C, and 5hmC remains undeaminated and is read as C. Consequently, the proportion of T readouts (P T) reflects the collective occurrences of C and 5mC, regulated by the deamination rate of 5mC (R 5mC). By determining R 5mC and P T values, we can deduce the precise levels of C, 5mC, and 5hmC at particular genomic locations. We successfully used EDA-seq to simultaneously measure C, 5mC, and 5hmC at specific loci within human lung cancer tissue and their normal counterpart. The results from EDA-seq demonstrated a strong concordance with those obtained from the combined application of BS-seq and ACE-seq methods. EDA-seq eliminates the need for bisulfite treatment, DNA oxidation or glycosylation and uniquely enables simultaneous quantification of C, 5mC and 5hmC at the same genomic locations.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

