A novel dual-release scaffold for fluorescent labels improves cyclic immunofluorescence
- PMID: 38966675
- PMCID: PMC11221530
- DOI: 10.1039/d4cb00007b
A novel dual-release scaffold for fluorescent labels improves cyclic immunofluorescence
Abstract
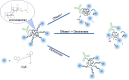
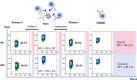
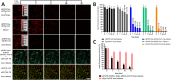
Cyclic immunofluorescence is a powerful method to generate high-content imaging datasets for investigating cell biology and developing therapies. This method relies on fluorescent labels that determine the quality of immunofluorescence and the maximum number of staining cycles that can be performed. Here we present a novel fluorescent labelling strategy, based on antibodies conjugated to a scaffold containing two distinct sites for enzymatic cleavage of fluorophores. The scaffold is composed of a dextran decorated with short ssDNA that upon hybridization with complementary dye-modified oligos result in fluorescent molecules. The developed fluorescent labels exhibit specific staining and remarkable brightness in flow cytometry and fluorescence microscopy. We showed that the combination of DNase-mediated degradation of DNA and dextranse-mediated degradation of the dextran as two complementary enzymatic release mechanisms in one molecule, improves signal erasure from labelled epitopes. We envision that such dual-release labels with high brightness and efficient and specific erasure will advance multiplexed cyclic immunofluorescence approaches and thereby will contribute to gaining new insights in cell biology.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare the following competing interests. T. R., C. D. and D. A. Y. declare the filing of a patent application (EP 4 177 607 A1: Bright and Releasable Labels for Cell Staining Based on the Conjugates with Several Sites of Fluorophore Release), which was assigned to Miltenyi Biotec B.V. & Co. KG.
Figures


References
-
- Joshi S. and Yu D., in Basic Science Methods for Clinical Researchers, ed., M. Jalali, F. Y. L. Saldanha and M. Jalali, Academic Press, Boston, 2017, pp. 135–150
-
- Reiber T. Zavoiura O. Dose C. Yushchenko D. A. Eur. J. Org. Chem. 2021:2817–2830. doi: 10.1002/ejoc.202100117. - DOI
LinkOut - more resources
Full Text Sources

