Data-Independent Acquisition Represents a Promising Alternative for Fast Photochemical Oxidation of Proteins (FPOP) Samples Analysis
- PMID: 38967040
- PMCID: PMC11256011
- DOI: 10.1021/acs.analchem.4c01084
Data-Independent Acquisition Represents a Promising Alternative for Fast Photochemical Oxidation of Proteins (FPOP) Samples Analysis
Abstract
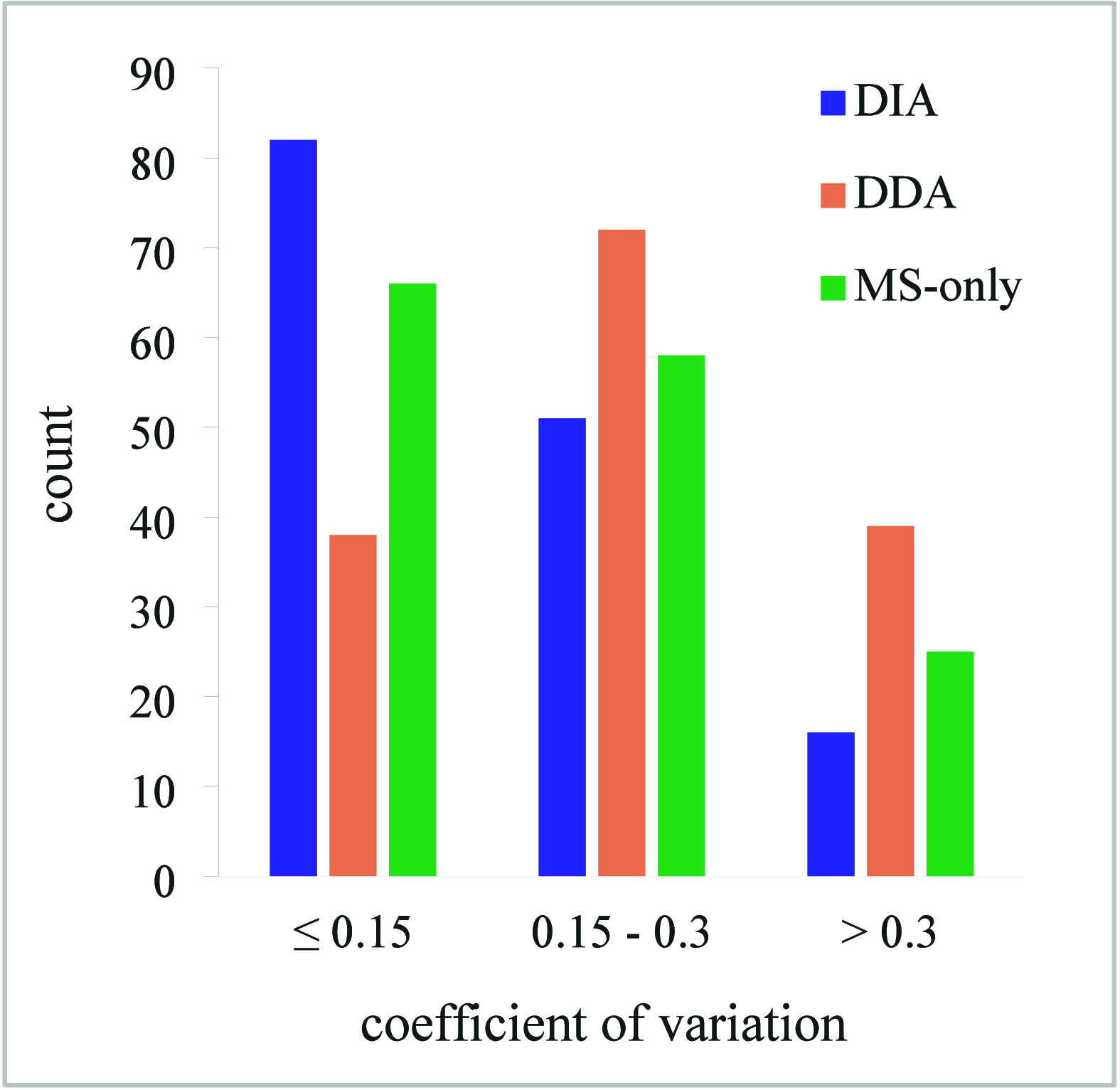
Fast Photochemical Oxidation of Proteins (FPOP) is a protein footprinting method utilizing hydroxyl radicals to provide valuable information on the solvent-accessible surface area. The extensive number of oxidative modifications that are created by FPOP is both advantageous, leading to great spatial resolution, and challenging, increasing the complexity of data processing. The precise localization of the modification together with the appropriate reproducibility is crucial to obtain relevant structural information. In this paper, we propose a novel approach combining validated spectral libraries together with utilizing DIA data. First, the DDA data searched by FragPipe are subsequently validated using Skyline software to form a spectral library. This library is then matched against the DIA data to filter out nonrepresentative IDs. In comparison with FPOP data processing using only a search engine followed by generally applied filtration steps, the manually validated spectral library offers higher confidence in identifications and increased spatial resolution. Furthermore, the reproducibility of quantification was compared for DIA, DDA, and MS-only acquisition modes on timsTOF SCP. Comparison of coefficients of variation (CV) showed that the DIA and MS acquisition modes exhibit significantly better reproducibility in quantification (CV medians 0.1233 and 0.1494, respectively) compared to the DDA mode (CV median 0.2104).
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization.Acc Chem Res. 2018 Mar 20;51(3):736-744. doi: 10.1021/acs.accounts.7b00593. Epub 2018 Feb 16. Acc Chem Res. 2018. PMID: 29450991 Free PMC article. Review.
-
Fast Photochemical Oxidation of Proteins Coupled with Mass Spectrometry.Protein Pept Lett. 2019;26(1):27-34. doi: 10.2174/0929866526666181128124554. Protein Pept Lett. 2019. PMID: 30484399 Free PMC article. Review.
-
Reproducibility, Specificity and Accuracy of Relative Quantification Using Spectral Library-based Data-independent Acquisition.Mol Cell Proteomics. 2020 Jan;19(1):181-197. doi: 10.1074/mcp.RA119.001714. Epub 2019 Nov 7. Mol Cell Proteomics. 2020. PMID: 31699904 Free PMC article.
-
Modifications generated by fast photochemical oxidation of proteins reflect the native conformations of proteins.Protein Sci. 2018 Jun;27(6):1047-1056. doi: 10.1002/pro.3408. Epub 2018 Apr 14. Protein Sci. 2018. PMID: 29575296 Free PMC article.
-
Fast photochemical oxidation of proteins (FPOP): A powerful mass spectrometry-based structural proteomics tool.J Biol Chem. 2019 Aug 9;294(32):11969-11979. doi: 10.1074/jbc.REV119.006218. Epub 2019 Jul 1. J Biol Chem. 2019. PMID: 31262727 Free PMC article. Review.
References
-
- Du Y.; Duc N. M.; Rasmussen S. G. F.; Hilger D.; Kubiak X.; Wang L.; Bohon J.; Kim H. R.; Wegrecki M.; Asuru A.; Jeong K. M.; Lee J.; Chance M. R.; Lodowski D. T.; Kobilka B. K.; Chung K. Y. Assembly of a GPCR-G Protein Complex. Cell 2019, 177 (5), 1232–1242.e11. 10.1016/j.cell.2019.04.022. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

