Orsay Virus Infection in Caenorhabditis elegans
- PMID: 38967546
- PMCID: PMC11486333
- DOI: 10.1002/cpz1.1098
Orsay Virus Infection in Caenorhabditis elegans
Abstract
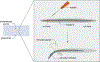
Orsay virus infection in the nematode Caenorhabditis elegans presents an opportunity to study host-virus interactions in an easily culturable, whole-animal host. Previously, a major limitation of C. elegans as a model for studying antiviral immunity was the lack of viruses known to naturally infect the worm. With the 2011 discovery of the Orsay virus, a naturally occurring viral pathogen, C. elegans has emerged as a compelling model for research on antiviral defense. From the perspective of the host, the genetic tractability of C. elegans enables mechanistic studies of antiviral immunity while the transparency of this animal allows for the observation of subcellular processes in vivo. Preparing infective virus filtrate and performing infections can be achieved with relative ease in a laboratory setting. Moreover, several tools are available to measure the outcome of infection. Here, we describe workflows for generating infective virus filtrate, achieving reproducible infection of C. elegans, and assessing the outcome of viral infection using molecular biology approaches and immunofluorescence. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Preparation of Orsay virus filtrate Support Protocol: Synchronize C. elegans development by bleaching Basic Protocol 2: Orsay virus infection Basic Protocol 3: Quantification of Orsay virus RNA1/RNA2 transcript levels by qRT-PCR Basic Protocol 4: Quantification of infection rate and fluorescence in situ hybridization (FISH) fluorescence intensity Basic Protocol 5: Immunofluorescent labeling of dsRNA in virus-infected intestinal tissue.
Keywords: C. elegans; Orsay virus; anti‐dsRNA immunofluorescence; virus filtrate preparation; virus infection.
© 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Figures




Similar articles
-
A targeted genetic screen identifies Caenorhabditis elegans genes involved in RNAi-independent antiviral defense.Virology. 2025 Sep;610:110597. doi: 10.1016/j.virol.2025.110597. Epub 2025 Jun 11. Virology. 2025. PMID: 40516488
-
Decreased SynMuv B gene activity in response to viral infection leads to activation of the antiviral RNAi pathway in C. elegans.PLoS Biol. 2025 Jan 29;23(1):e3002748. doi: 10.1371/journal.pbio.3002748. eCollection 2025 Jan. PLoS Biol. 2025. PMID: 39879188 Free PMC article.
-
Story of an infection: Viral dynamics and host responses in the Caenorhabditis elegans-Orsay virus pathosystem.Sci Adv. 2024 Sep 27;10(39):eadn5945. doi: 10.1126/sciadv.adn5945. Epub 2024 Sep 27. Sci Adv. 2024. PMID: 39331715 Free PMC article.
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
Cited by
-
Adenosine deaminase and deoxyadenosine regulate intracellular immune response in C. elegans.iScience. 2025 Feb 3;28(3):111950. doi: 10.1016/j.isci.2025.111950. eCollection 2025 Mar 21. iScience. 2025. PMID: 40034845 Free PMC article.
-
Conserved chromatin regulators control the transcriptional immune response to intracellular pathogens in Caenorhabditis elegans.PLoS Genet. 2025 Apr 7;21(4):e1011444. doi: 10.1371/journal.pgen.1011444. eCollection 2025 Apr. PLoS Genet. 2025. PMID: 40193347 Free PMC article.
References
-
- Batachari LE, Dai AY, and Troemel ER 2024. C. elegans RIG-I-like receptor DRH-1 signals via CARDs to activate anti-viral immunity in intestinal cells. 2024.02.05.578694. Available at: https://www.biorxiv.org/content/10.1101/2024.02.05.578694v1 [Accessed April 14, 2024]. - DOI - PMC - PubMed
-
- Corsi AK, Wightman B, and Chalfie M 2018. A Transparent window into biology: A primer on Caenorhabditis elegans. In WormBook: The Online Review of C. elegans Biology [Internet] WormBook Available at: https://www.ncbi.nlm.nih.gov/books/NBK299460/ [Accessed March 26, 2024]. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

