Competition and synergy of Arp2/3 and formins in nucleating actin waves
- PMID: 38968072
- PMCID: PMC11378572
- DOI: 10.1016/j.celrep.2024.114423
Competition and synergy of Arp2/3 and formins in nucleating actin waves
Abstract
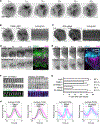
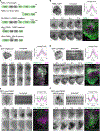
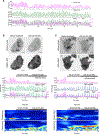
Actin assembly and dynamics are crucial for maintaining cell structure and changing physiological states. The broad impact of actin on various cellular processes makes it challenging to dissect the specific role of actin regulatory proteins. Using actin waves that propagate on the cortex of mast cells as a model, we discovered that formins (FMNL1 and mDia3) are recruited before the Arp2/3 complex in actin waves. GTPase Cdc42 interactions drive FMNL1 oscillations, with active Cdc42 and the constitutively active mutant of FMNL1 capable of forming waves on the plasma membrane independently of actin waves. Additionally, the delayed recruitment of Arp2/3 antagonizes FMNL1 and active Cdc42. This antagonism is not due to competition for monomeric actin but rather for their common upstream regulator, active Cdc42, whose levels are negatively regulated by Arp2/3 via SHIP1 recruitment. Collectively, our study highlights the complex feedback loops in the dynamic control of the actin cytoskeletal network.
Keywords: CP: Cell biology; actin waves; dynamical systems; limited pool model.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
Competition and Synergy of Arp2/3 and Formins in Nucleating Actin Waves.bioRxiv [Preprint]. 2023 Sep 13:2023.09.13.557508. doi: 10.1101/2023.09.13.557508. bioRxiv. 2023. Update in: Cell Rep. 2024 Jul 23;43(7):114423. doi: 10.1016/j.celrep.2024.114423. PMID: 37745345 Free PMC article. Updated. Preprint.
Similar articles
-
Competition and Synergy of Arp2/3 and Formins in Nucleating Actin Waves.bioRxiv [Preprint]. 2023 Sep 13:2023.09.13.557508. doi: 10.1101/2023.09.13.557508. bioRxiv. 2023. Update in: Cell Rep. 2024 Jul 23;43(7):114423. doi: 10.1016/j.celrep.2024.114423. PMID: 37745345 Free PMC article. Updated. Preprint.
-
Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex.Curr Biol. 2007 Apr 3;17(7):579-91. doi: 10.1016/j.cub.2007.03.024. Curr Biol. 2007. PMID: 17398099
-
Capping protein is dispensable for polarized actin network growth and actin-based motility.J Biol Chem. 2020 Nov 6;295(45):15366-15375. doi: 10.1074/jbc.RA120.015009. Epub 2020 Aug 31. J Biol Chem. 2020. PMID: 32868296 Free PMC article.
-
Actin polymerization downstream of integrins: signaling pathways and mechanotransduction.Biochem J. 2020 Jan 17;477(1):1-21. doi: 10.1042/BCJ20170719. Biochem J. 2020. PMID: 31913455 Review.
-
Formins as effector proteins of Rho GTPases.Small GTPases. 2014;5:e29513. doi: 10.4161/sgtp.29513. Epub 2014 Jun 10. Small GTPases. 2014. PMID: 24914801 Free PMC article. Review.
Cited by
-
FGF-Mediated Axon Guidance: Role of Downstream Signaling Pathways in Cytoskeletal Control.Cells. 2025 May 25;14(11):777. doi: 10.3390/cells14110777. Cells. 2025. PMID: 40497953 Free PMC article. Review.
-
Synthetic Forms Most Beautiful: Engineering Insights into Self-Organization.Physiology (Bethesda). 2025 Jul 1;40(4):0. doi: 10.1152/physiol.00064.2024. Epub 2025 Feb 12. Physiology (Bethesda). 2025. PMID: 39938118 Free PMC article. Review.
-
Collective dynamics of actin and microtubule and its crosstalk mediated by FHDC1.Front Cell Dev Biol. 2024 Mar 19;11:1261117. doi: 10.3389/fcell.2023.1261117. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38567385 Free PMC article.
-
An atypical Arp2/3 complex is required for Plasmodium DNA segregation and malaria transmission.Nat Microbiol. 2025 Jul;10(7):1775-1790. doi: 10.1038/s41564-025-02023-6. Epub 2025 Jun 13. Nat Microbiol. 2025. PMID: 40514561 Free PMC article.
-
Self-organizing glycolytic waves tune cellular metabolic states and fuel cancer progression.Nat Commun. 2025 Jul 1;16(1):5563. doi: 10.1038/s41467-025-60596-6. Nat Commun. 2025. PMID: 40593532 Free PMC article.
References
-
- Lappalainen P, Kotila T, Jégou A, and Romet-Lemonne G. (2022). Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol 23, 836–852. - PubMed
-
- Chhabra ES, and Higgs HN (2007). The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol 9, 1110–1121. - PubMed
-
- Vicker MG (2002). Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp. Cell Res 275, 54–66. - PubMed
-
- Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Müller-Taubenberger A, Köhler J, and Gerisch G. (2004). Dynamic Actin Patterns and Arp2/3 Assembly at the Substrate-Attached Surface of Motile Cells. Curr. Biol 14, 1–10. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

