SARS-CoV-2 pathogenesis in an angiotensin II-induced heart-on-a-chip disease model and extracellular vesicle screening
- PMID: 38968108
- PMCID: PMC11253010
- DOI: 10.1073/pnas.2403581121
SARS-CoV-2 pathogenesis in an angiotensin II-induced heart-on-a-chip disease model and extracellular vesicle screening
Abstract
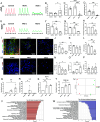
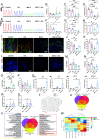
Adverse cardiac outcomes in COVID-19 patients, particularly those with preexisting cardiac disease, motivate the development of human cell-based organ-on-a-chip models to recapitulate cardiac injury and dysfunction and for screening of cardioprotective therapeutics. Here, we developed a heart-on-a-chip model to study the pathogenesis of SARS-CoV-2 in healthy myocardium established from human induced pluripotent stem cell (iPSC)-derived cardiomyocytes and a cardiac dysfunction model, mimicking aspects of preexisting hypertensive disease induced by angiotensin II (Ang II). We recapitulated cytopathic features of SARS-CoV-2-induced cardiac damage, including progressively impaired contractile function and calcium handling, apoptosis, and sarcomere disarray. SARS-CoV-2 presence in Ang II-treated hearts-on-a-chip decreased contractile force with earlier onset of contractile dysfunction and profoundly enhanced inflammatory cytokines compared to SARS-CoV-2 alone. Toward the development of potential therapeutics, we evaluated the cardioprotective effects of extracellular vesicles (EVs) from human iPSC which alleviated the impairment of contractile force, decreased apoptosis, reduced the disruption of sarcomeric proteins, and enhanced beta-oxidation gene expression. Viral load was not affected by either Ang II or EV treatment. We identified MicroRNAs miR-20a-5p and miR-19a-3p as potential mediators of cardioprotective effects of these EVs.
Keywords: SARS-CoV-2; cardiomyocyte; induced pluripotent stem cell; myocardium; organ-on-a-chip.
Conflict of interest statement
Competing interests statement:Y.Z., G.V.-N., B.Z., and M.R. are inventors on patents for cardiac tissue cultivation that are licensed to Valo Health. Q.W., Y.Z., and M.R. have a filed patent application on thermoplastic polymer composition for micro 3D printing and uses thereof. B.Z. holds equity in OrganoBiotech.
Figures




References
MeSH terms
Substances
Grants and funding
- 2R01 HL076485/HHS | NIH (NIH)
- FDN-167274/Canadian Government | Canadian Institutes of Health Research (CIHR)
- 19119 and 36442/Canada Foundation for Innovation/Ontario Research Fund
- RGPIN 326982-10/Natural Sciences and Engineering Research Council of Canada Discovery Grant
- R01 HL076485/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

