Single-cell genomics details the maturation block in BCP-ALL and identifies therapeutic vulnerabilities in DUX4-r cases
- PMID: 38968149
- PMCID: PMC11451301
- DOI: 10.1182/blood.2023021705
Single-cell genomics details the maturation block in BCP-ALL and identifies therapeutic vulnerabilities in DUX4-r cases
Abstract
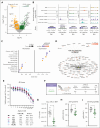
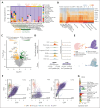
B-cell progenitor acute lymphoblastic leukemia (BCP-ALL) is the most common childhood malignancy and is driven by multiple genetic alterations that cause maturation arrest and accumulation of abnormal progenitor B cells. Current treatment protocols with chemotherapy have led to favorable outcomes but are associated with significant toxicity and risk of side effects, highlighting the necessity for highly effective, less toxic, targeted drugs, even in subtypes with a favorable outcome. Here, we used multimodal single-cell sequencing to delineate the transcriptional, epigenetic, and immunophenotypic characteristics of 23 childhood BCP-ALLs belonging to the BCR::ABL1+, ETV6::RUNX1+, high hyperdiploid, and recently discovered DUX4-rearranged (DUX4-r) subtypes. Projection of the ALL cells along the normal hematopoietic differentiation axis revealed a diversity in the maturation pattern between the different BCP-ALL subtypes. Although the BCR::ABL1+, ETV6::RUNX1+, and high hyperdiploidy cells mainly showed similarities to normal pro-B cells, DUX4-r ALL cells also displayed transcriptional signatures resembling mature B cells. Focusing on the DUX4-r subtype, we found that the blast population displayed not only multilineage priming toward nonhematopoietic cells, myeloid, and T-cell lineages, but also an activation of phosphatidylinositol 3-kinase (PI3K)/AKT signaling that sensitized the cells to PI3K inhibition in vivo. Given the multilineage priming of DUX4-r blasts with aberrant expression of myeloid marker CD371 (CLL-1), we generated chimeric antigen receptor T cells, which effectively eliminated DUX4-r ALL cells in vivo. These results provide a detailed characterization of BCP-ALL at the single-cell level and reveal therapeutic vulnerabilities in the DUX4-r subtype, with implications for the understanding of ALL biology and new therapeutic strategies.
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures





Comment in
-
Targeting DUX4-r leukemia via PI3K and CD371.Blood. 2024 Sep 26;144(13):1354-1356. doi: 10.1182/blood.2024025988. Blood. 2024. PMID: 39325481 No abstract available.
References
-
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–1548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

