Monitoring emerging pathogens using negative nucleic acid test results from endemic pathogens in pig populations: Application to porcine enteric coronaviruses
- PMID: 38968319
- PMCID: PMC11226029
- DOI: 10.1371/journal.pone.0306532
Monitoring emerging pathogens using negative nucleic acid test results from endemic pathogens in pig populations: Application to porcine enteric coronaviruses
Abstract
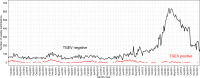
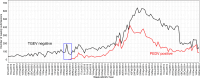
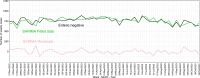
This study evaluated the use of endemic enteric coronaviruses polymerase chain reaction (PCR)-negative testing results as an alternative approach to detect the emergence of animal health threats with similar clinical diseases presentation. This retrospective study, conducted in the United States, used PCR-negative testing results from porcine samples tested at six veterinary diagnostic laboratories. As a proof of concept, the database was first searched for transmissible gastroenteritis virus (TGEV) negative submissions between January 1st, 2010, through April 29th, 2013, when the first porcine epidemic diarrhea virus (PEDV) case was diagnosed. Secondly, TGEV- and PEDV-negative submissions were used to detect the porcine delta coronavirus (PDCoV) emergence in 2014. Lastly, encountered best detection algorithms were implemented to prospectively monitor the 2023 enteric coronavirus-negative submissions. Time series (weekly TGEV-negative counts) and Seasonal Autoregressive-Integrated Moving-Average (SARIMA) were used to control for outliers, trends, and seasonality. The SARIMA's fitted and residuals were then subjected to anomaly detection algorithms (EARS, EWMA, CUSUM, Farrington) to identify alarms, defined as weeks of higher TGEV-negativity than what was predicted by models preceding the PEDV emergence. The best-performing detection algorithms had the lowest false alarms (number of alarms detected during the baseline) and highest time to detect (number of weeks between the first alarm and PEDV emergence). The best-performing detection algorithms were CUSUM, EWMA, and Farrington flexible using SARIMA fitted values, having a lower false alarm rate and identified alarms 4 to 17 weeks before PEDV and PDCoV emergences. No alarms were identified in the 2023 enteric negative testing results. The negative-based monitoring system functioned in the case of PEDV propagating epidemic and in the presence of a concurrent propagating epidemic with the PDCoV emergence. It demonstrated its applicability as an additional tool for diagnostic data monitoring of emergent pathogens having similar clinical disease as the monitored endemic pathogens.
Copyright: © 2024 Serafini Poeta Silva et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Dórea FC, Sanchez J, Revie CW. Veterinary syndromic surveillance: Current initiatives and potential for development. Prev Vet Med. 2011. Aug;101(1–2):1–17. - PubMed
-
- Santman-Berends IMGA, Brouwer-Middelesch H, Van Wuijckhuise L, de Bont-Smolenaars AJG, Van Schaik G. Surveillance of cattle health in the Netherlands: Monitoring trends and developments using routinely collected cattle census data. Prev Vet Med. 2016. Nov;134:103–12. doi: 10.1016/j.prevetmed.2016.10.002 - DOI - PubMed
-
- Trevisan G, Linhares LCM, Crim B, Dubey P, Schwartz KJ, Burrough ER, et al.. Macroepidemiological aspects of porcine reproductive and respiratory syndrome virus detection by major United States veterinary diagnostic laboratories over time, age group, and specimen. PLoS One. 2019. Oct 16;14(10):e0223544. doi: 10.1371/journal.pone.0223544 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

