mRNA-encoded Cas13 treatment of Influenza via site-specific degradation of genomic RNA
- PMID: 38968329
- PMCID: PMC11253931
- DOI: 10.1371/journal.ppat.1012345
mRNA-encoded Cas13 treatment of Influenza via site-specific degradation of genomic RNA
Abstract
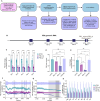
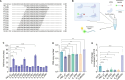
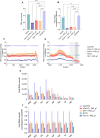
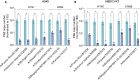
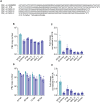
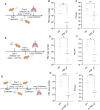
The CRISPR-Cas13 system has been proposed as an alternative treatment of viral infections. However, for this approach to be adopted as an antiviral, it must be optimized until levels of efficacy rival or exceed the performance of conventional approaches. To take steps toward this goal, we evaluated the influenza viral RNA degradation patterns resulting from the binding and enzymatic activity of mRNA-encoded LbuCas13a and two crRNAs from a prior study, targeting PB2 genomic and messenger RNA. We found that the genome targeting guide has the potential for significantly higher potency than originally detected, because degradation of the genomic RNA is not uniform across the PB2 segment, but it is augmented in proximity to the Cas13 binding site. The PB2 genome targeting guide exhibited high levels (>1 log) of RNA degradation when delivered 24 hours post-infection in vitro and maintained that level of degradation over time, with increasing multiplicity of infection (MOI), and across modern influenza H1N1 and H3N2 strains. Chemical modifications to guides with potent LbuCas13a function, resulted in nebulizer delivered efficacy (>1-2 log reduction in viral titer) in a hamster model of influenza (Influenza A/H1N1/California/04/09) infection given prophylactically or as a treatment (post-infection). Maximum efficacy was achieved with two doses, when administered both pre- and post-infection. This work provides evidence that mRNA-encoded Cas13a can effectively mitigate Influenza A infections opening the door to the development of a programmable approach to treating multiple respiratory infections.
Copyright: © 2024 Chaves et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
D.V. and P.J.S. have a patent applied for regarding the polymer used in this work. D.V., C.Z. and P.J.S. have a provisional patent filed for the guides described in this work, and are co-founders of Tether Therapeutics. The terms of these arrangements have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies. All other authors declare no competing interests.
Figures

References
-
- Grohskopf LA, Blanton LH, Ferdinands JM, Chung JR, Broder KR, Talbot HK, et al. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices—United States, 2022–23 Influenza Season. MMWR Recomm Rep. 2022;71: 1–28. doi: 10.15585/mmwr.rr7101a1 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

