Neural progenitor-derived Apelin controls tip cell behavior and vascular patterning
- PMID: 38968355
- PMCID: PMC11225789
- DOI: 10.1126/sciadv.adk1174
Neural progenitor-derived Apelin controls tip cell behavior and vascular patterning
Abstract
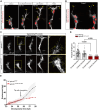
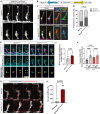
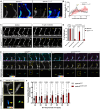
During angiogenesis, vascular tip cells guide nascent vascular sprouts to form a vascular network. Apelin, an agonist of the G protein-coupled receptor Aplnr, is enriched in vascular tip cells, and it is hypothesized that vascular-derived Apelin regulates sprouting angiogenesis. We identify an apelin-expressing neural progenitor cell population in the dorsal neural tube. Vascular tip cells exhibit directed elongation and migration toward and along the apelin-expressing neural progenitor cells. Notably, restoration of neural but not vascular apelin expression in apelin mutants remedies the angiogenic defects of mutants. By functional analyses, we show the requirement of Apelin signaling for tip cell behaviors, like filopodia formation and cell elongation. Through genetic interaction studies and analysis of transgenic activity reporters, we identify Apelin signaling as a modulator of phosphoinositide 3-kinase and extracellular signal-regulated kinase signaling in tip cells in vivo. Our results suggest a previously unidentified neurovascular cross-talk mediated by Apelin signaling that is important for tip cell function during sprouting angiogenesis.
Figures





References
-
- Jakobsson L., Franco C. A., Bentley K., Collins R. T., Ponsioen B., Aspalter I. M., Rosewell I., Busse M., Thurston G., Medvinsky A., Schulte-Merker S., Gerhardt H., Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat. Cell Biol. 12, 943–953 (2010). - PubMed
-
- Phng L. K., Stanchi F., Gerhardt H., Filopodia are dispensable for endothelial tip cell guidance. Development 140, 4031–4040 (2013). - PubMed
-
- Bussmann J., Bos F. L., Urasaki A., Kawakami K., Duckers H. J., Schulte-Merker S., Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development 137, 2653–2657 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

