Prospective, international, multisite comparison of platelet isolation techniques for genome-wide transcriptomics: communication from the SSC of the ISTH
- PMID: 38969303
- PMCID: PMC11416310
- DOI: 10.1016/j.jtha.2024.06.017
Prospective, international, multisite comparison of platelet isolation techniques for genome-wide transcriptomics: communication from the SSC of the ISTH
Abstract
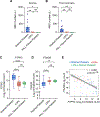
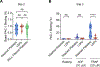
Genome-wide platelet transcriptomics is increasingly used to uncover new aspects of platelet biology and as a diagnostic and prognostic tool. Nevertheless, platelet isolation methods for transcriptomic studies are not standardized, introducing challenges for cross-study comparisons, data integration, and replication. In this prospective multicenter study, called "Standardizing Platelet Transcriptomics for Discovery, Diagnostics, and Therapeutics in the Thrombosis and Hemostasis Community (STRIDE)" by the International Society on Thrombosis and Haemostasis Scientific and Standardization Committees, we assessed how 3 of the most commonly used platelet isolation protocols influence metrics from next-generation bulk RNA sequencing and functional assays. Compared with washing alone, more stringent removal of leukocytes by anti-CD45 beads or PALL filters resulted in a sufficient quantity of RNA for next-generation sequencing and similar quality of RNA sequencing metrics. Importantly, stringent removal of leukocytes resulted in the lower relative expression of known leukocyte-specific genes and the higher relative expression of known platelet-specific genes. The results were consistent across enrolling sites, suggesting that the techniques are transferrable and reproducible. Moreover, all 3 isolation techniques did not influence basal platelet reactivity, but agonist-induced integrin αIIbβ3 activation is reduced by anti-CD45 bead isolation compared with washing alone. In conclusion, the isolation technique chosen influences genome-wide transcriptional and functional assays in platelets. These results should help the research community make informed choices about platelet isolation techniques in their own platelet studies.
Keywords: leukocytes; next-generation RNA-seq; platelet transcriptomics; platelets.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interests M.T.R. reports patents pending or issued on using platelet transcriptomics. All other authors have no conflicts to declare.
Figures
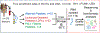



References
-
- Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA, Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118(14):e101–11. Epub 20110519. doi: 10.1182/blood-2011-03-339705. PubMed PMID: 21596849; PMCID: PMC3193274. - DOI - PMC - PubMed
-
- Supernat A, Popeda M, Pastuszak K, Best MG, Gresner P, Veld SI, Siek B, Bednarz-Knoll N, Rondina MT, Stokowy T, Wurdinger T, Jassem J, Zaczek AJ. Transcriptomic landscape of blood platelets in healthy donors. Sci Rep. 2021;11(1):15679. Epub 20210803. doi: 10.1038/s41598-021-94003-z. PubMed PMID: 34344933; PMCID: PMC8333095. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

