Exploring and retrieving sequence and metadata for species across the tree of life with NCBI Datasets
- PMID: 38969627
- PMCID: PMC11226681
- DOI: 10.1038/s41597-024-03571-y
Exploring and retrieving sequence and metadata for species across the tree of life with NCBI Datasets
Abstract
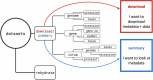
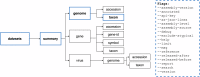
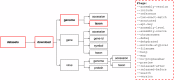
To explore complex biological questions, it is often necessary to access various data types from public data repositories. As the volume and complexity of biological sequence data grow, public repositories face significant challenges in ensuring that the data is easily discoverable and usable by the biological research community. To address these challenges, the National Center for Biotechnology Information (NCBI) has created NCBI Datasets. This resource provides straightforward, comprehensive, and scalable access to biological sequences, annotations, and metadata for a wide range of taxa. Following the FAIR (Findable, Accessible, Interoperable, and Reusable) data management principles, NCBI Datasets offers user-friendly web interfaces, command-line tools, and documented APIs, empowering researchers to access NCBI data seamlessly. The data is delivered as packages of sequences and metadata, thus facilitating improved data retrieval, sharing, and usability in research. Moreover, this data delivery method fosters effective data attribution and promotes its further reuse. This paper outlines the current scope of data accessible through NCBI Datasets and explains various options for exploring and downloading the data.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lathe W, Williams J, Mangan M, Karolchik D. Genomic Data Resources: Challenges and Promises. Nature Education. 2008;1(3):2.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

