CNS-wide repopulation by hematopoietic-derived microglia-like cells corrects progranulin deficiency in mice
- PMID: 38969669
- PMCID: PMC11226701
- DOI: 10.1038/s41467-024-49908-4
CNS-wide repopulation by hematopoietic-derived microglia-like cells corrects progranulin deficiency in mice
Abstract
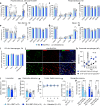
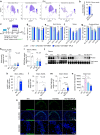
Hematopoietic stem cell transplantation can deliver therapeutic proteins to the central nervous system (CNS) through transplant-derived microglia-like cells. However, current conditioning approaches result in low and slow engraftment of transplanted cells in the CNS. Here we optimized a brain conditioning regimen that leads to rapid, robust, and persistent microglia replacement without adverse effects on neurobehavior or hematopoiesis. This regimen combines busulfan myeloablation and six days of Colony-stimulating factor 1 receptor inhibitor PLX3397. Single-cell analyses revealed unappreciated heterogeneity of microglia-like cells with most cells expressing genes characteristic of homeostatic microglia, brain-border-associated macrophages, and unique markers. Cytokine analysis in the CNS showed transient inductions of myeloproliferative and chemoattractant cytokines that help repopulate the microglia niche. Bone marrow transplant of progranulin-deficient mice conditioned with busulfan and PLX3397 restored progranulin in the brain and eyes and normalized brain lipofuscin storage, proteostasis, and lipid metabolism. This study advances our understanding of CNS repopulation by hematopoietic-derived cells and demonstrates its therapeutic potential for treating progranulin-dependent neurodegeneration.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Update of
-
CNS Repopulation by Hematopoietic-Derived Microglia-Like Cells Corrects Progranulin deficiency.Res Sq [Preprint]. 2023 Sep 12:rs.3.rs-3263412. doi: 10.21203/rs.3.rs-3263412/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Jul 5;15(1):5654. doi: 10.1038/s41467-024-49908-4. PMID: 37790525 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

