Functional multi-organelle units control inflammatory lipid metabolism of macrophages
- PMID: 38969763
- PMCID: PMC11321999
- DOI: 10.1038/s41556-024-01457-0
Functional multi-organelle units control inflammatory lipid metabolism of macrophages
Abstract
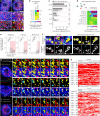
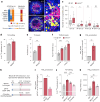
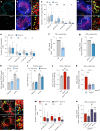
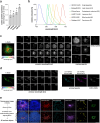
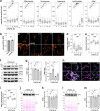
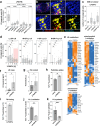
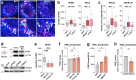
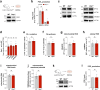
Eukaryotic cells contain several membrane-separated organelles to compartmentalize distinct metabolic reactions. However, it has remained unclear how these organelle systems are coordinated when cells adapt metabolic pathways to support their development, survival or effector functions. Here we present OrgaPlexing, a multi-spectral organelle imaging approach for the comprehensive mapping of six key metabolic organelles and their interactions. We use this analysis on macrophages, immune cells that undergo rapid metabolic switches upon sensing bacterial and inflammatory stimuli. Our results identify lipid droplets (LDs) as primary inflammatory responder organelle, which forms three- and four-way interactions with other organelles. While clusters with endoplasmic reticulum (ER) and mitochondria (mitochondria-ER-LD unit) help supply fatty acids for LD growth, the additional recruitment of peroxisomes (mitochondria-ER-peroxisome-LD unit) supports fatty acid efflux from LDs. Interference with individual components of these units has direct functional consequences for inflammatory lipid mediator synthesis. Together, we show that macrophages form functional multi-organellar units to support metabolic adaptation and provide an experimental strategy to identify organelle-metabolic signalling hubs.
© 2024. The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

