An improved 3D-UNet-based brain hippocampus segmentation model based on MR images
- PMID: 38970025
- PMCID: PMC11225132
- DOI: 10.1186/s12880-024-01346-w
An improved 3D-UNet-based brain hippocampus segmentation model based on MR images
Abstract
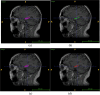
Objective: Accurate delineation of the hippocampal region via magnetic resonance imaging (MRI) is crucial for the prevention and early diagnosis of neurosystemic diseases. Determining how to accurately and quickly delineate the hippocampus from MRI results has become a serious issue. In this study, a pixel-level semantic segmentation method using 3D-UNet is proposed to realize the automatic segmentation of the brain hippocampus from MRI results.
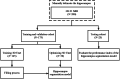
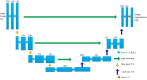
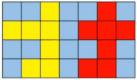
Methods: Two hundred three-dimensional T1-weighted (3D-T1) nongadolinium contrast-enhanced magnetic resonance (MR) images were acquired at Hangzhou Cancer Hospital from June 2020 to December 2022. These samples were divided into two groups, containing 175 and 25 samples. In the first group, 145 cases were used to train the hippocampus segmentation model, and the remaining 30 cases were used to fine-tune the hyperparameters of the model. Images for twenty-five patients in the second group were used as the test set to evaluate the performance of the model. The training set of images was processed via rotation, scaling, grey value augmentation and transformation with a smooth dense deformation field for both image data and ground truth labels. A filling technique was introduced into the segmentation network to establish the hippocampus segmentation model. In addition, the performance of models established with the original network, such as VNet, SegResNet, UNetR and 3D-UNet, was compared with that of models constructed by combining the filling technique with the original segmentation network.
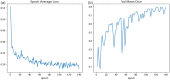
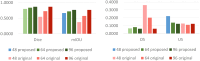
Results: The results showed that the performance of the segmentation model improved after the filling technique was introduced. Specifically, when the filling technique was introduced into VNet, SegResNet, 3D-UNet and UNetR, the segmentation performance of the models trained with an input image size of 48 × 48 × 48 improved. Among them, the 3D-UNet-based model with the filling technique achieved the best performance, with a Dice score (Dice score) of 0.7989 ± 0.0398 and a mean intersection over union (mIoU) of 0.6669 ± 0.0540, which were greater than those of the original 3D-UNet-based model. In addition, the oversegmentation ratio (OSR), average surface distance (ASD) and Hausdorff distance (HD) were 0.0666 ± 0.0351, 0.5733 ± 0.1018 and 5.1235 ± 1.4397, respectively, which were better than those of the other models. In addition, when the size of the input image was set to 48 × 48 × 48, 64 × 64 × 64 and 96 × 96 × 96, the model performance gradually improved, and the Dice scores of the proposed model reached 0.7989 ± 0.0398, 0.8371 ± 0.0254 and 0.8674 ± 0.0257, respectively. In addition, the mIoUs reached 0.6669 ± 0.0540, 0.7207 ± 0.0370 and 0.7668 ± 0.0392, respectively.
Conclusion: The proposed hippocampus segmentation model constructed by introducing the filling technique into a segmentation network performed better than models built solely on the original network and can improve the efficiency of diagnostic analysis.
Keywords: 3D-UNet; Brain hippocampus segmentation; Deep learning; Filling technique; MRI.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
[Fully Automatic Glioma Segmentation Algorithm of Magnetic Resonance Imaging Based on 3D-UNet With More Global Contextual Feature Extraction: An Improvement on Insufficient Extraction of Global Features].Sichuan Da Xue Xue Bao Yi Xue Ban. 2024 Mar 20;55(2):447-454. doi: 10.12182/20240360208. Sichuan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38645864 Free PMC article. Chinese.
-
Deep learning-based recognition and segmentation of intracranial aneurysms under small sample size.Front Physiol. 2022 Dec 19;13:1084202. doi: 10.3389/fphys.2022.1084202. eCollection 2022. Front Physiol. 2022. PMID: 36601346 Free PMC article.
-
A benchmark study of convolutional neural networks in fully automatic segmentation of aortic root.Front Bioeng Biotechnol. 2023 Jun 15;11:1171868. doi: 10.3389/fbioe.2023.1171868. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37397959 Free PMC article.
-
Label-driven magnetic resonance imaging (MRI)-transrectal ultrasound (TRUS) registration using weakly supervised learning for MRI-guided prostate radiotherapy.Phys Med Biol. 2020 Jun 26;65(13):135002. doi: 10.1088/1361-6560/ab8cd6. Phys Med Biol. 2020. PMID: 32330922 Free PMC article.
-
Attention-UNet architectures with pretrained backbones for multi-class cardiac MR image segmentation.Curr Probl Cardiol. 2024 Jan;49(1 Pt C):102129. doi: 10.1016/j.cpcardiol.2023.102129. Epub 2023 Oct 20. Curr Probl Cardiol. 2024. PMID: 37866419 Review.
References
-
- Adult Central. Nervous System Tumors Treatment (PDQ®)–Health Professional Version. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

