miR-455/GREM1 axis promotes colorectal cancer progression and liver metastasis by affecting PI3K/AKT pathway and inducing M2 macrophage polarization
- PMID: 38970064
- PMCID: PMC11225248
- DOI: 10.1186/s12935-024-03422-1
miR-455/GREM1 axis promotes colorectal cancer progression and liver metastasis by affecting PI3K/AKT pathway and inducing M2 macrophage polarization
Abstract
Background: Colorectal cancer is among the most common malignant tumors affecting the gastrointestinal tract. Liver metastases, a complication present in approximately 50% of colorectal cancer patients, are a considerable concern. Recently, studies have revealed the crucial role of miR-455 in tumor pathogenesis. However, the effect of miR-455 on the progression of liver metastases in colorectal cancer remains controversial. As an antagonist of bone morphogenetic protein(BMP), Gremlin 1 (GREM1) may impact organogenesis, body patterning, and tissue differentiation. Nevertheless, the role of miR-455 in regulating GREM1 in colorectal cancer liver metastases and how miR-455/GREM1 axis influences tumour immune microenvironment is unclear.
Methods: Bioinformatics analysis shows that miR-455/GREM1 axis plays crucial role in liver metastasis of intestinal cancer and predicts its possible mechanism. To investigate the impact of miR-455/GREM1 axis on the proliferation, invasion, and migration of colorectal cancer cells, colony formation assay, wound healing and transwell assay were examined in vitro. The Dual-Luciferase reporter gene assay and RNA pull-down assay confirmed a possible regulatory effect between miR-455 and GREM1. In vivo, colorectal cancer liver metastasis(CRLM) model mice was established to inquiry the effect of miR-455/GREM1 axis on tumor growth and macrophage polarization. The marker of macrophage polarization was tested using immunofluorescence(IF) and quantitative real-time polymerase chain reaction(qRT-PCR). By enzyme-linked immunosorbent assay (ELISA), cytokines were detected in culture medium supernatants.
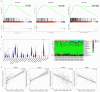
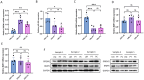
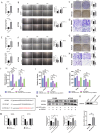
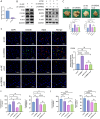
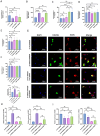
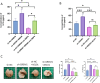
Results: We found that miR-455 and BMP6 expression was increased and GREM1 expression was decreased in liver metastase compared with primary tumor. miR-455/GREM1 axis promotes colorectal cancer cells proliferation, migration, invasion via affected PI3K/AKT pathway. Moreover, downregulating GREM1 augmented BMP6 expression in MC38 cell lines, inducing M2 polarization of macrophages, and promoting liver metastasis growth in CRLM model mice.
Conclusion: These data suggest that miR-455/GREM1 axis promotes colorectal cancer progression and liver metastasis by affecting PI3K/AKT pathway and inducing M2 macrophage polarization. These results offer valuable insights and direction for future research and treatment of CRLM.
Keywords: AKT; Colorectal cancer; GREM1; Liver metastasis; Macrophage polarization; PI3K; miR-455.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

