Metatranscriptomics-guided genome-scale metabolic reconstruction reveals the carbon flux and trophic interaction in methanogenic communities
- PMID: 38970122
- PMCID: PMC11225162
- DOI: 10.1186/s40168-024-01830-z
Metatranscriptomics-guided genome-scale metabolic reconstruction reveals the carbon flux and trophic interaction in methanogenic communities
Abstract
Background: Despite rapid advances in genomic-resolved metagenomics and remarkable explosion of metagenome-assembled genomes (MAGs), the function of uncultivated anaerobic lineages and their interactions in carbon mineralization remain largely uncertain, which has profound implications in biotechnology and biogeochemistry.
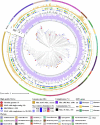
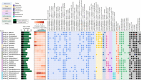
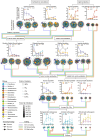
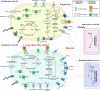
Results: In this study, we combined long-read sequencing and metatranscriptomics-guided metabolic reconstruction to provide a genome-wide perspective of carbon mineralization flow from polymers to methane in an anaerobic bioreactor. Our results showed that incorporating long reads resulted in a substantial improvement in the quality of metagenomic assemblies, enabling the effective recovery of 132 high-quality genomes meeting stringent criteria of minimum information about a metagenome-assembled genome (MIMAG). In addition, hybrid assembly obtained 51% more prokaryotic genes in comparison to the short-read-only assembly. Metatranscriptomics-guided metabolic reconstruction unveiled the remarkable metabolic flexibility of several novel Bacteroidales-affiliated bacteria and populations from Mesotoga sp. in scavenging amino acids and sugars. In addition to recovering two circular genomes of previously known but fragmented syntrophic bacteria, two newly identified bacteria within Syntrophales were found to be highly engaged in fatty acid oxidation through syntrophic relationships with dominant methanogens Methanoregulaceae bin.74 and Methanothrix sp. bin.206. The activity of bin.206 preferring acetate as substrate exceeded that of bin.74 with increasing loading, reinforcing the substrate determinantal role.
Conclusion: Overall, our study uncovered some key active anaerobic lineages and their metabolic functions in this complex anaerobic ecosystem, offering a framework for understanding carbon transformations in anaerobic digestion. These findings advance the understanding of metabolic activities and trophic interactions between anaerobic guilds, providing foundational insights into carbon flux within both engineered and natural ecosystems. Video Abstract.
Keywords: Anaerobic digestion; Long reads; Omics; Reverse electron transfer; Syntrophic bacteria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- IRENA. Renewable energy Statistics 2022. Abu Dhabi: International Renewable Energy Agency Abu Dhabi; 2022.
-
- Association WB. Global potential of biogas. London: World Biogas Association; 2019.
-
- Kamagata Y, Tamaki H. Cultivation of uncultured fastidious microbes. Microbes Environ. 2005;20:85–91. doi: 10.1264/jsme2.20.85. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

