A live cell imaging-based assay for tracking particle uptake by clathrin-mediated endocytosis
- PMID: 38971609
- PMCID: PMC11609598
- DOI: 10.1016/bs.mie.2024.02.010
A live cell imaging-based assay for tracking particle uptake by clathrin-mediated endocytosis
Abstract
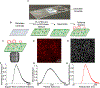
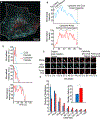
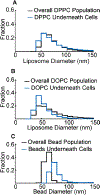
A popular strategy for therapeutic delivery to cells and tissues is to encapsulate therapeutics inside particles that cells internalize via endocytosis. The efficacy of particle uptake by endocytosis is often studied in bulk using flow cytometry and Western blot analysis and confirmed using confocal microscopy. However, these techniques do not reveal the detailed dynamics of particle internalization and how the inherent heterogeneity of many types of particles may impact their endocytic uptake. Toward addressing these gaps, here we present a live-cell imaging-based method that utilizes total internal reflection fluorescence microscopy to track the uptake of a large ensemble of individual particles in parallel, as they interact with the cellular endocytic machinery. To analyze the resulting data, we employ an open-source tracking algorithm in combination with custom data filters. This analysis reveals the dynamic interactions between particles and endocytic structures, which determine the probability of particle uptake. In particular, our approach can be used to examine how variations in the physical properties of particles (size, targeting, rigidity), as well as heterogeneity within the particle population, impact endocytic uptake. These data impact the design of particles toward more selective and efficient delivery of therapeutics to cells.
Keywords: Clathrin-mediated endocytosis; Colocalization; Drug-carrier internalization; Drug-carrier properties; Endocytosis; Particle tracking; Small unilamellar vesicle.
Copyright © 2024. Published by Elsevier Inc.
Figures




References
-
- Aguet F, Upadhyayula S, Gaudin R, Chou Y, Cocucci E, He K, Chen B-C, Mosaliganti K, Pasham M, Skillern W, Legant WR, Liu T-L, Findlay G, Marino E, Danuser G, Megason S, Betzig E, & Kirchhausen T (2016). Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Molecular Biology of the Cell, 27(22), 3418–3435. 10.1091/mbc.E16-03-0164 - DOI - PMC - PubMed
-
- Antonny B, Burd C, De Camilli P, Chen E, Daumke O, Faelber K, Ford M, Frolov VA, Frost A, Hinshaw JE, Kirchhausen T, Kozlov MM, Lenz M, Low HH, McMahon H, Merrifield C, Pollard TD, Robinson PJ, Roux A, & Schmid S (2016). Membrane fission by dynamin: What we know and what we need to know. The EMBO Journal, 35(21), 2270–2284. 10.15252/embj.201694613 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

