Development and validation of machine learning models to predict MDRO colonization or infection on ICU admission by using electronic health record data
- PMID: 38971777
- PMCID: PMC11227715
- DOI: 10.1186/s13756-024-01428-y
Development and validation of machine learning models to predict MDRO colonization or infection on ICU admission by using electronic health record data
Abstract
Background: Multidrug-resistant organisms (MDRO) pose a significant threat to public health. Intensive Care Units (ICU), characterized by the extensive use of antimicrobial agents and a high prevalence of bacterial resistance, are hotspots for MDRO proliferation. Timely identification of patients at high risk for MDRO can aid in curbing transmission, enhancing patient outcomes, and maintaining the cleanliness of the ICU environment. This study focused on developing a machine learning (ML) model to identify patients at risk of MDRO during the initial phase of their ICU stay.
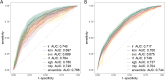
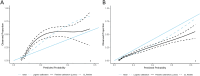
Methods: Utilizing patient data from the First Medical Center of the People's Liberation Army General Hospital (PLAGH-ICU) and the Medical Information Mart for Intensive Care (MIMIC-IV), the study analyzed variables within 24 h of ICU admission. Machine learning algorithms were applied to these datasets, emphasizing the early detection of MDRO colonization or infection. Model efficacy was evaluated by the area under the receiver operating characteristics curve (AUROC), alongside internal and external validation sets.
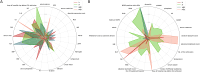
Results: The study evaluated 3,536 patients in PLAGH-ICU and 34,923 in MIMIC-IV, revealing MDRO prevalence of 11.96% and 8.81%, respectively. Significant differences in ICU and hospital stays, along with mortality rates, were observed between MDRO positive and negative patients. In the temporal validation, the PLAGH-ICU model achieved an AUROC of 0.786 [0.748, 0.825], while the MIMIC-IV model reached 0.744 [0.723, 0.766]. External validation demonstrated reduced model performance across different datasets. Key predictors included biochemical markers and the duration of pre-ICU hospital stay.
Conclusions: The ML models developed in this study demonstrated their capability in early identification of MDRO risks in ICU patients. Continuous refinement and validation in varied clinical contexts remain essential for future applications.
Keywords: Infection; Intensive care unit; Machine learning; Multidrug-resistant organisms; Predictive modeling.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. - DOI - PubMed
-
- Serra-Burriel M, Keys M, Campillo-Artero C, Agodi A, Barchitta M, Gikas A, Palos C, López-Casasnovas G. Impact of multi-drug resistant bacteria on economic and clinical outcomes of healthcare-associated infections in adults: systematic review and meta-analysis. PLoS ONE. 2020;15(1):e0227139. doi: 10.1371/journal.pone.0227139. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

