ATP6V0A1-dependent cholesterol absorption in colorectal cancer cells triggers immunosuppressive signaling to inactivate memory CD8+ T cells
- PMID: 38971819
- PMCID: PMC11227557
- DOI: 10.1038/s41467-024-50077-7
ATP6V0A1-dependent cholesterol absorption in colorectal cancer cells triggers immunosuppressive signaling to inactivate memory CD8+ T cells
Abstract
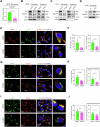
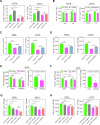
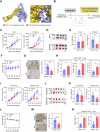
Obesity shapes anti-tumor immunity through lipid metabolism; however, the mechanisms underlying how colorectal cancer (CRC) cells utilize lipids to suppress anti-tumor immunity remain unclear. Here, we show that tumor cell-intrinsic ATP6V0A1 drives exogenous cholesterol-induced immunosuppression in CRC. ATP6V0A1 facilitates cholesterol absorption in CRC cells through RAB guanine nucleotide exchange factor 1 (RABGEF1)-dependent endosome maturation, leading to cholesterol accumulation within the endoplasmic reticulum and elevated production of 24-hydroxycholesterol (24-OHC). ATP6V0A1-induced 24-OHC upregulates TGF-β1 by activating the liver X receptor (LXR) signaling. Subsequently, the release of TGF-β1 into the tumor microenvironment by CRC cells activates the SMAD3 pathway in memory CD8+ T cells, ultimately suppressing their anti-tumor activities. Moreover, we identify daclatasvir, a clinically used anti-hepatitis C virus (HCV) drug, as an ATP6V0A1 inhibitor that can effectively enhance the memory CD8+ T cell activity and suppress tumor growth in CRC. These findings shed light on the potential for ATP6V0A1-targeted immunotherapy in CRC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

