Decoding cellular plasticity and niche regulation of limbal stem cells during corneal wound healing
- PMID: 38971839
- PMCID: PMC11227725
- DOI: 10.1186/s13287-024-03816-y
Decoding cellular plasticity and niche regulation of limbal stem cells during corneal wound healing
Abstract
Background: Dysfunction or deficiency of corneal epithelium results in vision impairment or blindness in severe cases. The rapid and effective regeneration of corneal epithelial cells relies on the limbal stem cells (LSCs). However, the molecular and functional responses of LSCs and their niche cells to injury remain elusive.
Methods: Single-cell RNA sequencing was performed on corneal tissues from normal mice and corneal epithelium defect models. Bioinformatics analysis was performed to confirm the distinct characteristics and cell fates of LSCs. Knockdown of Creb5 and OSM treatment experiment were performed to determine their roles of in corneal epithelial wound healing.
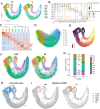
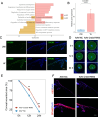
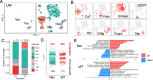
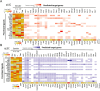
Results: Our data defined the molecular signatures of LSCs and reconstructed the pseudotime trajectory of corneal epithelial cells. Gene network analyses characterized transcriptional landmarks that potentially regulate LSC dynamics, and identified a transcription factor Creb5, that was expressed in LSCs and significantly upregulated after injury. Loss-of-function experiments revealed that silencing Creb5 delayed the corneal epithelial healing and LSC mobilization. Through cell-cell communication analysis, we identified 609 candidate regeneration-associated ligand-receptor interaction pairs between LSCs and distinct niche cells, and discovered a unique subset of Arg1+ macrophages infiltrated after injury, which were present as the source of Oncostatin M (OSM), an IL-6 family cytokine, that were demonstrated to effectively accelerate the corneal epithelial wound healing.
Conclusions: This research provides a valuable single-cell resource and reference for the discovery of mechanisms and potential clinical interventions aimed at ocular surface reconstruction.
Keywords: Cornea; Corneal epithelium; Limbal stem cells; Niche regulation; Single-cell RNA sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Compartmentalized 3D bioprinting of the limbal niche with distinct hPSC-LSC subpopulations for corneal disease modeling.Acta Biomater. 2025 Jul 1;201:187-197. doi: 10.1016/j.actbio.2025.05.068. Epub 2025 May 29. Acta Biomater. 2025. PMID: 40449707
-
Production and Limbal Lineage Commitment of Aniridia Patient-Derived Induced Pluripotent Stem Cells.Stem Cells. 2023 Dec 14;41(12):1133-1141. doi: 10.1093/stmcls/sxad067. Stem Cells. 2023. PMID: 37632794
-
A systematic review on the effects of ROCK inhibitors on proliferation and/or differentiation in human somatic stem cells: A hypothesis that ROCK inhibitors support corneal endothelial healing via acting on the limbal stem cell niche.Ocul Surf. 2023 Jan;27:16-29. doi: 10.1016/j.jtos.2022.12.008. Epub 2022 Dec 28. Ocul Surf. 2023. PMID: 36586668
-
TNF-α Suppression Attenuates Limbal Stem Cell Damage in Ocular Injury.Cornea. 2024 Dec 3;44(6):762-771. doi: 10.1097/ICO.0000000000003738. Cornea. 2024. PMID: 39626088 Free PMC article.
-
Cultivated Autologous Limbal Epithelial Cell Transplantation: A Review of Clinical Trials and Applications.Cornea. 2025 Apr 29;44(9):1071-1077. doi: 10.1097/ICO.0000000000003884. Cornea. 2025. PMID: 40315274 Review.
Cited by
-
Prox1 Protein in Corneal Limbal Lymphatic Vessels Maintains Limbal Stem Cell Stemness and Regulates Corneal Injury Repair.Invest Ophthalmol Vis Sci. 2025 Apr 1;66(4):81. doi: 10.1167/iovs.66.4.81. Invest Ophthalmol Vis Sci. 2025. PMID: 40298888 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

