An in-situ peptide-antibody self-assembly to block CD47 and CD24 signaling enhances macrophage-mediated phagocytosis and anti-tumor immune responses
- PMID: 38971872
- PMCID: PMC11227529
- DOI: 10.1038/s41467-024-49825-6
An in-situ peptide-antibody self-assembly to block CD47 and CD24 signaling enhances macrophage-mediated phagocytosis and anti-tumor immune responses
Abstract
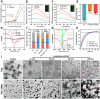
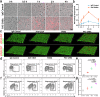
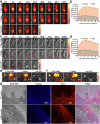
Targeted immunomodulation for reactivating innate cells, especially macrophages, holds great promise to complement current adaptive immunotherapy. Nevertheless, there is still a lack of high-performance therapeutics for blocking macrophage phagocytosis checkpoint inhibitors in solid tumors. Herein, a peptide-antibody combo-supramolecular in situ assembled CD47 and CD24 bi-target inhibitor (PAC-SABI) is described, which undergoes biomimetic surface propagation on cancer cell membranes through ligand-receptor binding and enzyme-triggered reactions. By simultaneously blocking CD47 and CD24 signaling, PAC-SABI enhances the phagocytic ability of macrophages in vitro and in vivo, promoting anti-tumor responses in breast and pancreatic cancer mouse models. Moreover, building on the foundation of PAC-SABI-induced macrophage repolarization and increased CD8+ T cell tumor infiltration, sequential anti-PD-1 therapy further suppresses 4T1 tumor progression, prolonging survival rate. The in vivo construction of PAC-SABI-based nano-architectonics provides an efficient platform for bridging innate and adaptive immunity to maximize therapeutic potency.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

