Crosstalk between RNA m6A modification and epigenetic factors in plant gene regulation
- PMID: 38971972
- PMCID: PMC11573915
- DOI: 10.1016/j.xplc.2024.101037
Crosstalk between RNA m6A modification and epigenetic factors in plant gene regulation
Abstract
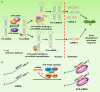
N6-methyladenosine (m6A) is the most abundant modification observed in eukaryotic mRNAs. Advances in transcriptome-wide m6A mapping and sequencing technologies have enabled the identification of several conserved motifs in plants, including the RRACH (R = A/G and H = A/C/U) and UGUAW (W = U or A) motifs. However, the mechanisms underlying deposition of m6A marks at specific positions in the conserved motifs of individual transcripts remain to be clarified. Evidence from plant and animal studies suggests that m6A writer or eraser components are recruited to specific genomic loci through interactions with particular transcription factors, 5-methylcytosine DNA methylation marks, and histone marks. In addition, recent studies in animal cells have shown that microRNAs play a role in depositing m6A marks at specific sites in transcripts through a base-pairing mechanism. m6A also affects the biogenesis and function of chromatin-associated regulatory RNAs and long noncoding RNAs. Although we have less of an understanding of the link between m6A modification and epigenetic factors in plants than in animals, recent progress in identifying the proteins that interact with m6A writer or eraser components has provided insights into the crosstalk between m6A modification and epigenetic factors, which plays a crucial role in transcript-specific methylation and regulation of m6A in plants.
Keywords: DNA methylation; RNA methylation; epigenetics; histone modification; long noncoding RNA; microRNA.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Reading m6A marks in mRNA: A potent mechanism of gene regulation in plants.J Integr Plant Biol. 2024 Dec;66(12):2586-2599. doi: 10.1111/jipb.13781. Epub 2024 Oct 4. J Integr Plant Biol. 2024. PMID: 39364713 Free PMC article. Review.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Covalent RNA modifications and their budding crosstalk with plant epigenetic processes.Curr Opin Plant Biol. 2022 Oct;69:102287. doi: 10.1016/j.pbi.2022.102287. Epub 2022 Aug 18. Curr Opin Plant Biol. 2022. PMID: 35988352 Review.
-
Crosstalk between histone/DNA modifications and RNA N6-methyladenosine modification.Curr Opin Genet Dev. 2024 Jun;86:102205. doi: 10.1016/j.gde.2024.102205. Epub 2024 May 21. Curr Opin Genet Dev. 2024. PMID: 38776766 Review.
-
Evolution and post-transcriptional regulation insights of m6A writers, erasers, and readers in plant epitranscriptome.Plant J. 2024 Oct;120(2):505-525. doi: 10.1111/tpj.16996. Epub 2024 Aug 21. Plant J. 2024. PMID: 39167634
Cited by
-
M6A Modified miR-31-5p Suppresses M1 Macrophage Polarization and Autoimmune Dry Eye by Targeting P2RX7.Adv Sci (Weinh). 2025 May;12(17):e2415341. doi: 10.1002/advs.202415341. Epub 2025 Mar 11. Adv Sci (Weinh). 2025. PMID: 40068094 Free PMC article.
-
Reading m6A marks in mRNA: A potent mechanism of gene regulation in plants.J Integr Plant Biol. 2024 Dec;66(12):2586-2599. doi: 10.1111/jipb.13781. Epub 2024 Oct 4. J Integr Plant Biol. 2024. PMID: 39364713 Free PMC article. Review.
-
Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation.Plants (Basel). 2025 Jun 30;14(13):2002. doi: 10.3390/plants14132002. Plants (Basel). 2025. PMID: 40648011 Free PMC article. Review.
-
RNA modifications in plant adaptation to abiotic stresses.Plant Commun. 2025 Feb 10;6(2):101229. doi: 10.1016/j.xplc.2024.101229. Epub 2024 Dec 21. Plant Commun. 2025. PMID: 39709520 Free PMC article. Review.
-
Non-coding RNA-mediated regulation of seed endosperm development.Front Plant Sci. 2025 Aug 8;16:1640284. doi: 10.3389/fpls.2025.1640284. eCollection 2025. Front Plant Sci. 2025. PMID: 40860731 Free PMC article. Review.
References
-
- Amara U., Hu J., Park S.J., Kang H. ECT12, an YTH-domain protein, is a potential mRNA m6A reader that affects abiotic stress responses by modulating mRNA stability in Arabidopsis. Plant Physiol. Biochem. 2024;206:108255. - PubMed
-
- Amara U., Shoaib Y., Kang H. ALKBH9C, a potential RNA m6A demethylase, regulates the response of Arabidopsis to abiotic stresses and abscisic acid. Plant Cell Environ. 2022;45:3566–3581. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

