Unveiling novel genetic variants in 370 challenging medically relevant genes using the long read sequencing data of 41 samples from 19 global populations
- PMID: 38972030
- PMCID: PMC11955097
- DOI: 10.1007/s00438-024-02158-x
Unveiling novel genetic variants in 370 challenging medically relevant genes using the long read sequencing data of 41 samples from 19 global populations
Abstract
Background: A large number of challenging medically relevant genes (CMRGs) are situated in complex or highly repetitive regions of the human genome, hindering comprehensive characterization of genetic variants using next-generation sequencing technologies. In this study, we employed long-read sequencing technology, extensively utilized in studying complex genomic regions, to characterize genetic alterations, including short variants (single nucleotide variants and short insertions and deletions) and copy number variations, in 370 CMRGs across 41 individuals from 19 global populations.
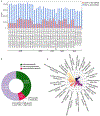
Results: Our analysis revealed high levels of genetic variants in CMRGs, with 68.73% exhibiting copy number variations and 65.20% containing short variants that may disrupt protein function across individuals. Such variants can influence pharmacogenomics, genetic disease susceptibility, and other clinical outcomes. We observed significant differences in CMRG variation across populations, with individuals of African ancestry harboring the highest number of copy number variants and short variants compared to samples from other continents. Notably, 15.79% to 33.96% of short variants were exclusively detectable through long-read sequencing. While the T2T-CHM13 reference genome significantly improved the assembly of CMRG regions, thereby facilitating variant detection in these regions, some regions still lacked resolution.
Conclusion: Our results provide an important reference for future clinical and pharmacogenetic studies, highlighting the need for a comprehensive representation of global genetic diversity in the reference genome and improved variant calling techniques to fully resolve medically relevant genes.
Keywords: Challenging medically relevant genes; Copy number variation; Genome sequencing; Long read sequencing; Short insertion and deletion; Single nucleotide polymorphism.
© 2024. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
Declarations
Figures
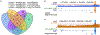


References
-
- Barile M, Giancaspero TA, Leone P et al. (2016) Riboflavin transport and metabolism in humans. J Inherit Metab Dis 39:545–557 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

