Identification and validation of a prognostic model based on four genes related to satellite nodules in hepatocellular carcinoma
- PMID: 38972883
- PMCID: PMC11228042
- DOI: 10.1038/s41598-024-66610-z
Identification and validation of a prognostic model based on four genes related to satellite nodules in hepatocellular carcinoma
Abstract
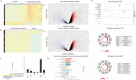
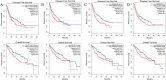
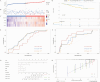
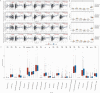
Satellite nodules is a key clinical characteristic which has prognostic value of hepatocellular carcinoma (HCC). Currently, there is no gene-level predictive model for Satellite nodules in liver cancer. For the 377 HCC cases collected from the dataset of Cancer Genome Atlas (TCGA), their original pathological data were analyzed to extract information regarding satellite nodules status as well as other relevant pathological data. Then, this study employed statistical modeling for prognostic model establishment in TCGA, and validation in International Cancer Genome Consortium (ICGC) cohorts and GSE76427. Through rigorous statistical analyses, 253 differential satellite nodules-related genes (SNRGs) were identified, and four key genes related to satellite nodules and prognosis were selected to construct a prognostic model. The high-risk group predicted by our model exhibited an unfavorable overall survival (OS) outlook and demonstrated an association with adverse worse clinical characteristics such as larger tumor size, higher alpha-fetoprotein, microvascular invasion and advanced stage. Moreover, the validation of the model's prognostic value in the ICGC and GSE76427 cohorts mirrored that of the TCGA cohort. Besides, the high-risk group also showed higher levels of resting Dendritic cells, M0 macrophages infiltration, alongside decreased levels of CD8+ T cells and γδT cells infiltration. The prognostic model based on SNRGs can reliability predict the OS of HCC and is likely to have predictive value of immunotherapy for HCC.
Keywords: Hepatocellular carcinoma; Prognostic model; Satellite nodules; Satellite nodules-related genes; Tumor-infiltrating lymphocytes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

