A deep learning-driven discovery of berberine derivatives as novel antibacterial against multidrug-resistant Helicobacter pylori
- PMID: 38972904
- PMCID: PMC11228022
- DOI: 10.1038/s41392-024-01895-0
A deep learning-driven discovery of berberine derivatives as novel antibacterial against multidrug-resistant Helicobacter pylori
Abstract
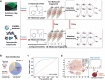
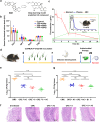
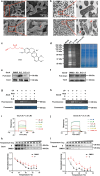
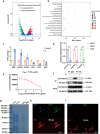
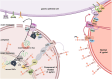
Helicobacter pylori (H. pylori) is currently recognized as the primary carcinogenic pathogen associated with gastric tumorigenesis, and its high prevalence and resistance make it difficult to tackle. A graph neural network-based deep learning model, employing different training sets of 13,638 molecules for pre-training and fine-tuning, was aided in predicting and exploring novel molecules against H. pylori. A positively predicted novel berberine derivative 8 with 3,13-disubstituted alkene exhibited a potency against all tested drug-susceptible and resistant H. pylori strains with minimum inhibitory concentrations (MICs) of 0.25-0.5 μg/mL. Pharmacokinetic studies demonstrated an ideal gastric retention of 8, with the stomach concentration significantly higher than its MIC at 24 h post dose. Oral administration of 8 and omeprazole (OPZ) showed a comparable gastric bacterial reduction (2.2-log reduction) to the triple-therapy, namely OPZ + amoxicillin (AMX) + clarithromycin (CLA) without obvious disturbance on the intestinal flora. A combination of OPZ, AMX, CLA, and 8 could further decrease the bacteria load (2.8-log reduction). More importantly, the mono-therapy of 8 exhibited comparable eradication to both triple-therapy (OPZ + AMX + CLA) and quadruple-therapy (OPZ + AMX + CLA + bismuth citrate) groups. SecA and BamD, playing a major role in outer membrane protein (OMP) transport and assembling, were identified and verified as the direct targets of 8 by employing the chemoproteomics technique. In summary, by targeting the relatively conserved OMPs transport and assembling system, 8 has the potential to be developed as a novel anti-H. pylori candidate, especially for the eradication of drug-resistant strains.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Oppong P, Majumdar D, Atherton J, Bebb J. Helicobacter pylori infection and peptic ulcers. Medicine. 2015;43:215–222. doi: 10.1016/j.mpmed.2015.01.008. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- 32141003/National Natural Science Foundation of China (National Science Foundation of China)
- 61972384/National Natural Science Foundation of China (National Science Foundation of China)
- 82003575/National Natural Science Foundation of China (National Science Foundation of China)
- 32141003/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical

