Using a new analytic approach for genotyping and phenotyping chromosome 9p deletion syndrome
- PMID: 38972963
- PMCID: PMC11369271
- DOI: 10.1038/s41431-024-01667-y
Using a new analytic approach for genotyping and phenotyping chromosome 9p deletion syndrome
Abstract
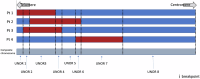
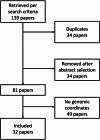
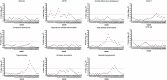
Using a new analytic method ("unique non-overlapping region" (UNOR) analysis), we characterized the genotypes and phenotypes of a large cohort of individuals diagnosed with chromosome 9p deletion syndrome (9PMS) and defined critical genomic regions. We extracted phenotypic information from 48 individuals with 9PMS from medical records and used a guided interview with caregivers to clarify ambiguities. Using high-resolution whole-genome sequencing for breakpoint definition, we aligned deletions and drew virtual breakpoints to obtain UNORs associated with phenotypic characteristics. We next extracted genotype and phenotype data for 57 individuals identified from a systematic review of the 9PMS literature and analyzed these as above. Common phenotypic features included developmental delay/intellectual disability, dysmorphic features, hypotonia, genital defects in XY individuals, psychiatric diagnoses, chronic constipation, atopic disease, vision problems, autism spectrum disorder, gastroesophageal reflux disease, trigonocephaly, congenital heart disease, and neonatal hypoglycemia. Our approach confirmed previous literature reports of an association of FREM1 with trigonocephaly and suggested a possible modifier element for this phenotype. In conclusion, the UNOR approach delineated phenotypic characteristics for 9PMS and confirmed the critical role of FREM1 and a possible long-distance regulatory element in pathogenesis of trigonocephaly that will need to be replicated in future studies.
© 2024. The Author(s), under exclusive licence to European Society of Human Genetics.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

