Genetic diversity of astroviruses detected in wild aquatic birds in Hong Kong
- PMID: 38972989
- PMCID: PMC11229208
- DOI: 10.1186/s12985-024-02423-w
Genetic diversity of astroviruses detected in wild aquatic birds in Hong Kong
Abstract
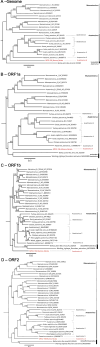
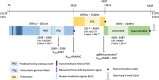
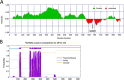
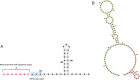
Wild waterfowl serve as a reservoir of some astroviruses. Fecal samples from wild waterfowl collected at Hong Kong's Marshes were tested using pan-astrovirus reverse transcription-PCR. Positive samples underwent subsequent host identification using DNA barcoding. Based on deduced partial sequences, noteworthy samples from three astrovirus groups (mammalian, avian and unclassified astroviruses) were further analyzed by next-generation sequencing. One sample of Avastrovirus 4 clade, MP22-196, had a nearly complete genome identified. The results of ORF2 phylogenetic analysis and genetic distance analysis indicate that Avastrovirus 4 is classified as a distinct subclade within Avastrovirus. MP22-196 has typical astrovirus genome characteristics. The unique characteristics and potential differences of this genome, compared to other avian astrovirus sequences, involve the identification of a modified sgRNA sequence situated near the ORF2 start codon, which precedes the ORF1b stop codon. Additionally, the 3' UTR of MP22-196 is shorter than other avian astroviruses. This study expands our understanding of the Avastrovirus 4 clade.
Keywords: Aquatic bird; Astrovirus; Avastrovirus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Knipe DM, Howley PM. Fields virology. 6th ed. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2013. 2 volumes p.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

