Protein biomarker signature in patients with spinal and bulbar muscular atrophy
- PMID: 38973610
- PMCID: PMC11383357
- DOI: 10.1172/jci.insight.176383
Protein biomarker signature in patients with spinal and bulbar muscular atrophy
Abstract
Spinal and bulbar muscular atrophy (SBMA) is a slowly progressing disease with limited sensitive biomarkers that support clinical research. We analyzed plasma and serum samples from patients with SBMA and matched healthy controls in multiple cohorts, identifying 40 highly reproducible SBMA-associated proteins out of nearly 3,000 measured. These proteins were robustly enriched in gene sets of skeletal muscle expression and processes related to mitochondria and calcium signaling. Many proteins outperformed currently used clinical laboratory tests (e.g., creatine kinase [CK]) in distinguishing patients from controls and in their correlations with clinical and functional traits in patients. Two of the 40 proteins, Ectodysplasin A2 receptor (EDA2R) and Repulsive guidance molecule A (RGMA), were found to be associated with decreased survival and body weight in a mouse model of SBMA. In summary, we identified what we believe to be a robust and novel set of fluid protein biomarkers in SBMA that are linked with relevant disease features in patients and in a mouse model of disease. Changes in these SBMA-associated proteins could be used as an early predictor of treatment effects in clinical trials.
Keywords: Genetic diseases; Muscle biology; Neuromuscular disease; Neuroscience; Proteomics.
Figures

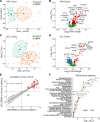
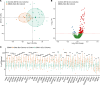
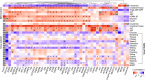

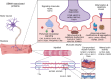
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

