IFN-λ uniquely promotes CD8 T cell immunity against SARS-CoV-2 relative to type I IFN
- PMID: 38973611
- PMCID: PMC11383353
- DOI: 10.1172/jci.insight.171830
IFN-λ uniquely promotes CD8 T cell immunity against SARS-CoV-2 relative to type I IFN
Abstract
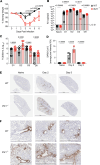
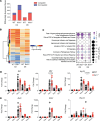
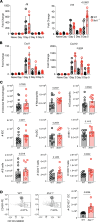
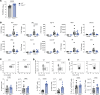
Optimization of protective immune responses against SARS-CoV-2 remains an urgent worldwide priority. In this regard, type III IFN (IFN-λ) restricts SARS-CoV-2 infection in vitro, and treatment with IFN-λ limits infection, inflammation, and pathogenesis in murine models. Furthermore, IFN-λ has been developed for clinical use to limit COVID-19 severity. However, whether endogenous IFN-λ signaling has an effect on SARS-CoV-2 antiviral immunity and long-term immune protection in vivo is unknown. In this study, we identified a requirement for IFN-λ signaling in promoting viral clearance and protective immune programming in SARS-CoV-2 infection of mice. Expression of both IFN and IFN-stimulated gene (ISG) in the lungs were minimally affected by the absence of IFN-λ signaling and correlated with transient increases in viral titers. We found that IFN-λ supported the generation of protective CD8 T cell responses against SARS-CoV-2 by facilitating accumulation of CD103+ DC in lung draining lymph nodes (dLN). IFN-λ signaling specifically in DCs promoted the upregulation of costimulatory molecules and the proliferation of CD8 T cells. Intriguingly, antigen-specific CD8 T cell immunity to SARS-CoV-2 was independent of type I IFN signaling, revealing a nonredundant function of IFN-λ. Overall, these studies demonstrate a critical role for IFN-λ in protective innate and adaptive immunity upon infection with SARS-CoV-2 and suggest that IFN-λ serves as an immune adjuvant to support CD8 T cell immunity.
Keywords: Immunology; Infectious disease; Innate immunity; Mouse models; T cells.
Conflict of interest statement
Figures


Similar articles
-
Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures.J Virol. 2020 Sep 15;94(19):e00985-20. doi: 10.1128/JVI.00985-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32699094 Free PMC article.
-
CCR2 Signaling Restricts SARS-CoV-2 Infection.mBio. 2021 Dec 21;12(6):e0274921. doi: 10.1128/mBio.02749-21. Epub 2021 Nov 9. mBio. 2021. PMID: 34749524 Free PMC article.
-
Differential immunoregulation by human surfactant protein A variants determines severity of SARS-CoV-2-induced lung disease.Front Immunol. 2025 Apr 2;16:1462278. doi: 10.3389/fimmu.2025.1462278. eCollection 2025. Front Immunol. 2025. PMID: 40242753 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
Workplace interventions to reduce the risk of SARS-CoV-2 infection outside of healthcare settings.Cochrane Database Syst Rev. 2022 May 6;5(5):CD015112. doi: 10.1002/14651858.CD015112.pub2. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 Apr 10;4:CD015112. doi: 10.1002/14651858.CD015112.pub3. PMID: 35514111 Free PMC article. Updated.
Cited by
-
Link of TMPRSS2 expression with tumor immunogenicity and response to immune checkpoint inhibitors in cancers.J Transl Med. 2025 Mar 7;23(1):294. doi: 10.1186/s12967-025-06177-z. J Transl Med. 2025. PMID: 40055791 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- 75N93022D00005/AI/NIAID NIH HHS/United States
- R01 HL168501/HL/NHLBI NIH HHS/United States
- R21 AI151230/AI/NIAID NIH HHS/United States
- 75N93023D00005/AI/NIAID NIH HHS/United States
- R35 GM150503/GM/NIGMS NIH HHS/United States
- R35 GM150806/GM/NIGMS NIH HHS/United States
- R21 AI174050/AI/NIAID NIH HHS/United States
- 75N95020D00005/DA/NIDA NIH HHS/United States
- R01 HL154001/HL/NHLBI NIH HHS/United States
- K22 AI146141/AI/NIAID NIH HHS/United States
- 75N99020D00005/OF/ORFDO NIH HHS/United States
- T32 AI165391/AI/NIAID NIH HHS/United States
- R01 AI130110/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

