Rapid and Amplification-free Nucleic Acid Detection with DNA Substrate-Mediated Autocatalysis of CRISPR/Cas12a
- PMID: 38973832
- PMCID: PMC11223203
- DOI: 10.1021/acsomega.4c03413
Rapid and Amplification-free Nucleic Acid Detection with DNA Substrate-Mediated Autocatalysis of CRISPR/Cas12a
Abstract
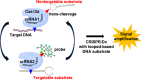
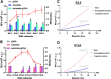
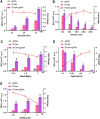
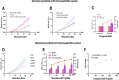
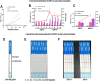
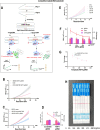
To enable rapid and accurate point-of-care DNA detection, we have developed a single-step, amplification-free nucleic acid detection platform, a DNA substrate-mediated autocatalysis of CRISPR/Cas12a (DSAC). DSAC makes use of the trans-cleavage activity of Cas12a and target template-activated DNA substrate for dual signal amplifications. DSAC employs two distinct DNA substrate types: one that enhances signal amplification and the other that negatively modulates fluorescent signals. The positive inducer utilizes nicked- or loop-based DNA substrates to activate CRISPR/Cas12a, initiating trans-cleavage activity in a positive feedback loop, ultimately amplifying the fluorescent signals. The negative modulator, which involves competitor-based DNA substrates, competes with the probes for trans-cleaving, resulting in a signal decline in the presence of target DNA. These DNA substrate-based DSAC systems were adapted to fluorescence-based and paper-based lateral flow strip detection platforms. Our DSAC system accurately detected African swine fever virus (ASFV) in swine's blood samples at femtomolar sensitivity within 20 min. In contrast to the existing amplification-free CRISPR/Dx platforms, DSAC offers a cost-effective and straightforward detection method, requiring only the addition of a rationally designed DNA oligonucleotide. Notably, a common ASFV sequence-encoded DNA substrate can be directly applied to detect human nucleic acids through a dual crRNA targeting system. Consequently, our single-step DSAC system presents an alternative point-of-care diagnostic tool for the sensitive, accurate, and timely diagnosis of viral infections with potential applicability to human disease detection.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Zetsche B.; Gootenberg J. S.; Abudayyeh O. O.; Slaymaker I. M.; Makarova K. S.; Essletzbichler P.; Volz S. E.; Joung J.; van der Oost J.; Regev A.; Koonin E. V.; Zhang F. Cpf1 Is a Single Rna-Guided Endonuclease of a Class 2 Crispr-Cas System. Cell 2015, 163, 759–771. 10.1016/j.cell.2015.09.038. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
