Comparative Genomic and Genetic Evidence on a Role for the OarX Protein in Thiamin Salvage
- PMID: 38973919
- PMCID: PMC11223231
- DOI: 10.1021/acsomega.4c03514
Comparative Genomic and Genetic Evidence on a Role for the OarX Protein in Thiamin Salvage
Abstract
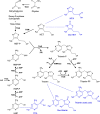
Salvage pathways for thiamin and its thiazole and pyrimidine moieties are poorly characterized compared to synthesis pathways. A candidate salvage gene is oarX, which encodes a short-chain dehydrogenase/reductase. In diverse bacteria, oarX clusters on the chromosome with genes of thiamin synthesis, salvage, or transport and is preceded by a thiamin pyrophosphate riboswitch. Thiamin and its moieties can undergo oxidations that convert a side-chain hydroxymethyl group to a carboxyl group, or the thiazole ring to a thiazolone, causing a loss of biological activity. To test if OarX participates in salvage of the carboxyl or thiazolone products, we used a genetic approach in Corynebacterium glutamicum ATCC 14067, which is auxotrophic for thiamin's pyrimidine moiety. This strain could not utilize the pyrimidine carboxyl derivative. This excluded a role in salvaging this product and narrowed the function search to metabolism of the carboxyl or thiazolone derivatives of thiamin or its thiazole moiety. However, a ΔthiG (thiazole auxotroph) strain was not rescued by any of these derivatives. Nor did deleting oarX affect rescue by the physiological pyrimidine and thiazole precursors of thiamin. These findings reinforce the genomic evidence that OarX has a function in thiamin metabolism and rule out five logical possibilities for what this function is.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Genomic insights into the thiamin metabolism of Paenibacillus thiaminolyticus NRRL B-4156 and P. apiarius NRRL B-23460.Stand Genomic Sci. 2017 Oct 3;12:59. doi: 10.1186/s40793-017-0276-9. eCollection 2017. Stand Genomic Sci. 2017. PMID: 29026451 Free PMC article.
-
The structural and biochemical foundations of thiamin biosynthesis.Annu Rev Biochem. 2009;78:569-603. doi: 10.1146/annurev.biochem.78.072407.102340. Annu Rev Biochem. 2009. PMID: 19348578 Free PMC article. Review.
-
Divisions of labor in the thiamin biosynthetic pathway among organs of maize.Front Plant Sci. 2014 Aug 4;5:370. doi: 10.3389/fpls.2014.00370. eCollection 2014. Front Plant Sci. 2014. PMID: 25136345 Free PMC article.
-
Identification of the thiamin salvage enzyme thiazole kinase in Arabidopsis and maize.Phytochemistry. 2013 Oct;94:68-73. doi: 10.1016/j.phytochem.2013.05.017. Epub 2013 Jun 28. Phytochemistry. 2013. PMID: 23816351
-
The genes and enzymes involved in the biosynthesis of thiamin and thiamin diphosphate in yeasts.Cell Mol Biol Lett. 2008;13(2):271-82. doi: 10.2478/s11658-007-0055-5. Epub 2008 Apr 10. Cell Mol Biol Lett. 2008. PMID: 18161008 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
