Bioinformatics analysis of an immunotherapy responsiveness-related gene signature in predicting lung adenocarcinoma prognosis
- PMID: 38973963
- PMCID: PMC11225057
- DOI: 10.21037/tlcr-24-309
Bioinformatics analysis of an immunotherapy responsiveness-related gene signature in predicting lung adenocarcinoma prognosis
Abstract
Background: Immune therapy has become first-line treatment option for patients with lung cancer, but some patients respond poorly to immune therapy, especially among patients with lung adenocarcinoma (LUAD). Novel tools are needed to screen potential responders to immune therapy in LUAD patients, to better predict the prognosis and guide clinical decision-making. Although many efforts have been made to predict the responsiveness of LUAD patients, the results were limited. During the era of immunotherapy, this study attempts to construct a novel prognostic model for LUAD by utilizing differentially expressed genes (DEGs) among patients with differential immune therapy responses.
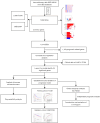
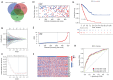
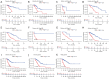
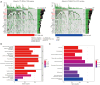
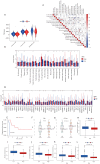
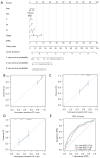
Methods: Transcriptome data of 598 patients with LUAD were downloaded from The Cancer Genome Atlas (TCGA) database, which included 539 tumor samples and 59 normal control samples, with a mean follow-up time of 29.69 months (63.1% of patients remained alive by the end of follow-up). Other data sources including three datasets from the Gene Expression Omnibus (GEO) database were analyzed, and the DEGs between immunotherapy responders and nonresponders were identified and screened. Univariate Cox regression analysis was applied with the TCGA cohort as the training set and GSE72094 cohort as the validation set, and least absolute shrinkage and selection operator (LASSO) Cox regression were applied in the prognostic-related genes which fulfilled the filter criteria to establish a prognostic formula, which was then tested with time-dependent receiver operating characteristic (ROC) analysis. Enriched pathways of the prognostic-related genes were analyzed with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, and tumor immune microenvironment (TIME), tumor mutational burden, and drug sensitivity tests were completed with appropriate packages in R (The R Foundation of Statistical Computing). Finally, a nomogram incorporating the prognostic formula was established.
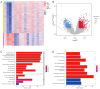
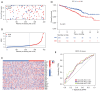
Results: A total of 1,636 DEGs were identified, 1,163 prognostic-related DEGs were extracted, and 34 DEGs were selected and incorporated into the immunotherapy responsiveness-related risk score (IRRS) formula. The IRRS formula had good performance in predicting the overall prognoses in patients with LUAD and had excellent performance in prognosis prediction in all LUAD subgroups. Moreover, the IRRS formula could predict anticancer drug sensitivity and immunotherapy responsiveness in patients with LUAD. Mechanistically, immune microenvironments varied profoundly between the two IRRS groups; the most significantly varied pathway between the high-IRRS and low-IRRS groups was ribonucleoprotein complex biogenesis, which correlated closely with the TP53 and TTN mutation burdens. In addition, we established a nomogram incorporating the IRRS, age, sex, clinical stage, T-stage, N-stage, and M-stage as predictors that could predict the prognoses of 1-year, 3-year, and 5-year survival in patients with LUAD, with an area under curve (AUC) of 0.718, 0.702, and 0.68, respectively.
Conclusions: The model we established in the present study could predict the prognosis of LUAD patients, help to identify patients with good responses to anticancer drugs and immunotherapy, and serve as a valuable tool to guide clinical decision-making.
Keywords: Lung adenocarcinoma (LUAD); anticancer drug sensitivity; immunotherapy responsiveness; nomogram; prognosis.
2024 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-309/coif). The authors have no conflicts of interest to declare.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
