Development of an ancestral DC and TLR4-inducing multi-epitope peptide vaccine against the spike protein of SARS-CoV and SARS-CoV-2 using the advanced immunoinformatics approaches
- PMID: 38974021
- PMCID: PMC11225186
- DOI: 10.1016/j.bbrep.2024.101745
Development of an ancestral DC and TLR4-inducing multi-epitope peptide vaccine against the spike protein of SARS-CoV and SARS-CoV-2 using the advanced immunoinformatics approaches
Abstract
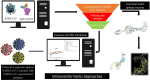
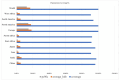
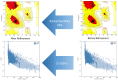
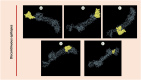
The oldest human coronavirus that started pandemics is severe acute respiratory syndrome virus (SARS-CoV). While SARS-CoV was eradicated, its new version, SARS-CoV2, caused the global pandemic of COVID-19. Evidence highlights the harmful events orchestrated by these viruses are mediated by Spike (S)P protein. Experimental epitopes of the S protein which were overlapping and ancestral between SARS-CoV and SARS-CoV-2 were obtained from the immune epitopes database (IEDB). The epitopes were then assembled in combination with a 50 S ribosomal protein L7/L12 adjuvant, a Mycobacterium tuberculosis-derived element and mediator of dendritic cells (DCs) and toll-like receptor 4 (TLR4). The immunogenic sequence was modeled by the GalaxyWeb server. After the improvement and validation of the protein structure, the physico-chemical properties and immune simulation were performed. To investigate the interaction with TLR3/4, Molecular Dynamics Simulation (MDS) was used. By merging the 17 B- and T-lymphocyte (HTL/CTL) epitopes, the vaccine sequence was created. Also, the Ramachandran plot presented that most of the residues were located in the most favorable and allowed areas. Moreover, SnapGene was successful in cloning the DNA sequence linked to our vaccine in the intended plasmid. A sequence was inserted between the XhoI and SacI position of the pET-28a (+) vector, and simulating the agarose gel revealed the existence of the inserted gene in the cloned plasmid with SARS vaccine (SARSV) construct, which has a 6565 bp in length overall. In terms of cytokines/IgG response, immunological simulation revealed a strong immune response. The stabilized vaccine showed strong interactions with TLR3/4, according to Molecular Dynamics Simulation (MDS) analysis. The present ancestral vaccine targets common sequences which seem to be valuable targets even for the new variant SARS-CoV-2.
Keywords: Computational immunology; Immunoinformatics; Multi-epitope; Reverse vaccinology; SARS-CoV; SARS-CoV-2.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures








Similar articles
-
Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2.Infect Dis Poverty. 2020 Sep 16;9(1):132. doi: 10.1186/s40249-020-00752-w. Infect Dis Poverty. 2020. PMID: 32938504 Free PMC article.
-
Applying high throughput and comprehensive immunoinformatics approaches to design a trivalent subunit vaccine for induction of immune response against emerging human coronaviruses SARS-CoV, MERS-CoV and SARS-CoV-2.J Biomol Struct Dyn. 2022 Aug;40(13):6097-6113. doi: 10.1080/07391102.2021.1876774. Epub 2021 Jan 29. J Biomol Struct Dyn. 2022. PMID: 33509045 Free PMC article.
-
Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach.J Biomol Struct Dyn. 2022 Jan;40(1):14-30. doi: 10.1080/07391102.2020.1792347. Epub 2020 Jul 17. J Biomol Struct Dyn. 2022. PMID: 32677533 Free PMC article.
-
Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches.Vaccine. 2020 Nov 10;38(48):7612-7628. doi: 10.1016/j.vaccine.2020.10.016. Epub 2020 Oct 9. Vaccine. 2020. PMID: 33082015 Free PMC article.
-
Multi-epitope vaccine against SARS-CoV-2 applying immunoinformatics and molecular dynamics simulation approaches.J Biomol Struct Dyn. 2022 Apr;40(7):2917-2933. doi: 10.1080/07391102.2020.1844060. Epub 2020 Nov 9. J Biomol Struct Dyn. 2022. PMID: 33164664 Free PMC article.
Cited by
-
Evaluating passive immunity in piglets from sows vaccinated with a PEDV S protein subunit vaccine.Front Cell Infect Microbiol. 2025 Jan 31;14:1498610. doi: 10.3389/fcimb.2024.1498610. eCollection 2024. Front Cell Infect Microbiol. 2025. PMID: 39963235 Free PMC article.
-
Interplay between innate-like T-cells and microRNAs in cancer immunity.Discov Oncol. 2025 Jul 28;16(1):1425. doi: 10.1007/s12672-025-03234-3. Discov Oncol. 2025. PMID: 40720066 Free PMC article. Review.
-
Bioinformatics-guided decoding of the Ancylostoma duodenale genome for the identification of potential vaccine targets.BMC Genomics. 2025 May 12;26(1):468. doi: 10.1186/s12864-025-11652-4. BMC Genomics. 2025. PMID: 40355819 Free PMC article.
-
Design of a novel multi-epitope vaccine candidate against Yersinia pestis using advanced immunoinformatics approaches: An in silico study.Biochem Biophys Rep. 2024 Nov 18;40:101871. doi: 10.1016/j.bbrep.2024.101871. eCollection 2024 Dec. Biochem Biophys Rep. 2024. PMID: 39634337 Free PMC article.
-
Multi-epitope vaccines: a promising strategy against viral diseases in swine.Front Cell Infect Microbiol. 2024 Dec 20;14:1497580. doi: 10.3389/fcimb.2024.1497580. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39760092 Free PMC article. Review.
References
-
- Loeb M.B. WHO; 2006. Severe Acute Respiratory Syndrome; pp. 465–472.https://www.who.int/health-topics/severe-acute-respiratory-syndrome#tab=... [Internet] Available from:
-
- Zeidler A., Karpinski T.M. SARS-CoV, MERS-CoV, SARS-CoV-2 comparison of three emerging coronaviruses. Jundishapur J. Microbiol. 2020;13(6) https://brieflands.com/articles/jjm-103744.html [Internet] Available from:
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

