Evaluation of a Covalent Library of Diverse Warheads (CovLib) Binding to JNK3, USP7, or p53
- PMID: 38974119
- PMCID: PMC11226190
- DOI: 10.2147/DDDT.S466829
Evaluation of a Covalent Library of Diverse Warheads (CovLib) Binding to JNK3, USP7, or p53
Abstract
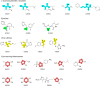
Purpose: Over the last few years, covalent fragment-based drug discovery has gained significant importance. Thus, striving for more warhead diversity, we conceived a library consisting of 20 covalently reacting compounds. Our covalent fragment library (CovLib) contains four different warhead classes, including five α-cyanoacacrylamides/acrylates (CA), three epoxides (EO), four vinyl sulfones (VS), and eight electron-deficient heteroarenes with a leaving group (SNAr/SN).
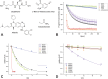
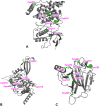
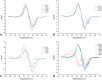
Methods: After predicting the theoretical solubility of the fragments by LogP and LogS during the selection process, we determined their experimental solubility using a turbidimetric solubility assay. The reactivities of the different compounds were measured in a high-throughput 5,5'-dithiobis-(2-nitrobenzoic acid) DTNB assay, followed by a (glutathione) GSH stability assay. We employed the CovLib in a (differential scanning fluorimetry) DSF-based screening against different targets: c-Jun N-terminal kinase 3 (JNK3), ubiquitin-specific protease 7 (USP7), and the tumor suppressor p53. Finally, the covalent binding was confirmed by intact protein mass spectrometry (MS).
Results: In general, the purchased fragments turned out to be sufficiently soluble. Additionally, they covered a broad spectrum of reactivity. All investigated α-cyanoacrylamides/acrylates and all structurally confirmed epoxides turned out to be less reactive compounds, possibly due to steric hindrance and reversibility (for α-cyanoacrylamides/acrylates). The SNAr and vinyl sulfone fragments are either highly reactive or stable. DSF measurements with the different targets JNK3, USP7, and p53 identified reactive fragment hits causing a shift in the melting temperatures of the proteins. MS confirmed the covalent binding mode of all these fragments to USP7 and p53, while additionally identifying the SNAr-type electrophile SN002 as a mildly reactive covalent hit for p53.
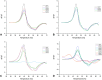
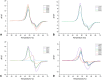
Conclusion: The screening and target evaluation of the CovLib revealed first interesting hits. The highly cysteine-reactive fragments VS004, SN001, SN006, and SN007 covalently modify several target proteins and showed distinct shifts in the melting temperatures up to +5.1 °C and -9.1 °C.
Keywords: 5,5'-dithiobis-(2-nitrobenzoic acid); covalent fragment-based drug discovery; differential scanning fluorimetry; glutathione; intact protein mass spectrometry.
© 2024 Klett et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

